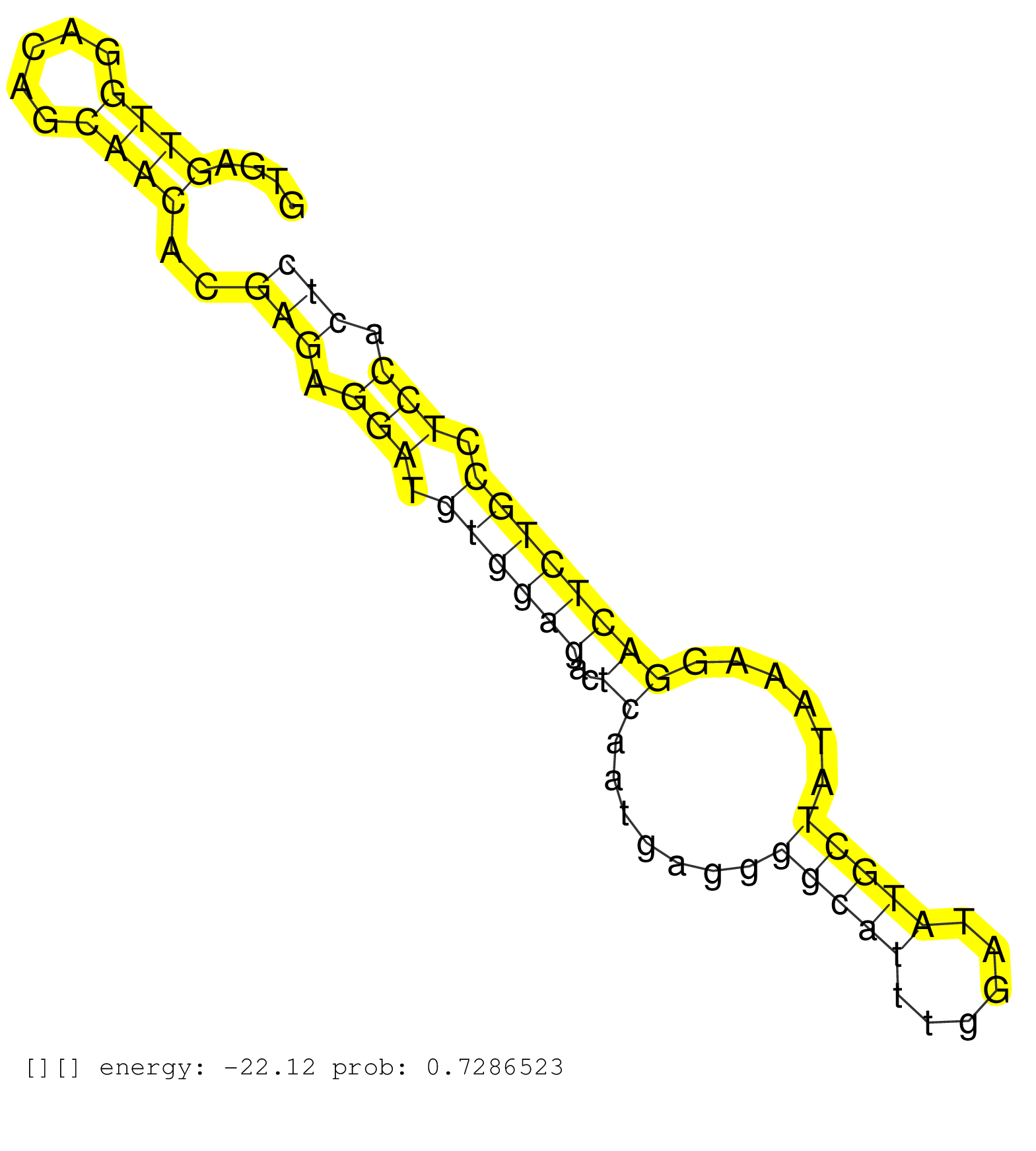

| Gene: Pnpla6 | ID: uc009krr.1_intron_9_0_chr8_3522669_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| TCCTGGCGCTTCACAATTACCTGGGTCTGACCAATGAACTCTTCAGTCATGTGAGTTGGACAGCAACACGAGAGGATGTGGAGACTCAATGAGGGGCATTTGGATATGCTATAAAGGACTCTGCCTCCACTCCACCACAGGAGATCCAGCCCCTACGCCTTTTCCCCAGCCCCGGCCTCCCGACCCGAAC ......................................................((((.....))))..(((.(((.((((((..((.......(((((......)))))......)))))))).))).))).......................................................... ..................................................51...............................................................................132........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTTGGACAGCAACACGAGAGGAT................................................................................................................. | 27 | 1 | 11.00 | 11.00 | 3.00 | 2.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ..................................................GTGAGTTGGACAGCAACACGAGAGGATG................................................................................................................ | 28 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGTTGGACAGCAACACGAGAGGATGT............................................................................................................... | 29 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................TGAGTTGGACAGCAACACGAGAGGATGT............................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGACAGCAACACGAGAGGA.................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................CTGACCAATGAACactc................................................................................................................................................... | 17 | actc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGACAGCAACACGAGAGGt.................................................................................................................. | 26 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CCTACGCCTTTTCCCC....................... | 16 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGACAGCAACACGAGAGGATt................................................................................................................ | 28 | t | 1.00 | 11.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGACAGCAACACGAGAGG................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................GATATGCTATAAAGGACTCTGCCTCC.............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| TCCTGGCGCTTCACAATTACCTGGGTCTGACCAATGAACTCTTCAGTCATGTGAGTTGGACAGCAACACGAGAGGATGTGGAGACTCAATGAGGGGCATTTGGATATGCTATAAAGGACTCTGCCTCCACTCCACCACAGGAGATCCAGCCCCTACGCCTTTTCCCCAGCCCCGGCCTCCCGACCCGAAC ......................................................((((.....))))..(((.(((.((((((..((.......(((((......)))))......)))))))).))).))).......................................................... ..................................................51...............................................................................132........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................TTTTCCCCAGCCCCGtt................ | 17 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCAGCCCCGGCCTCC.......... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - |