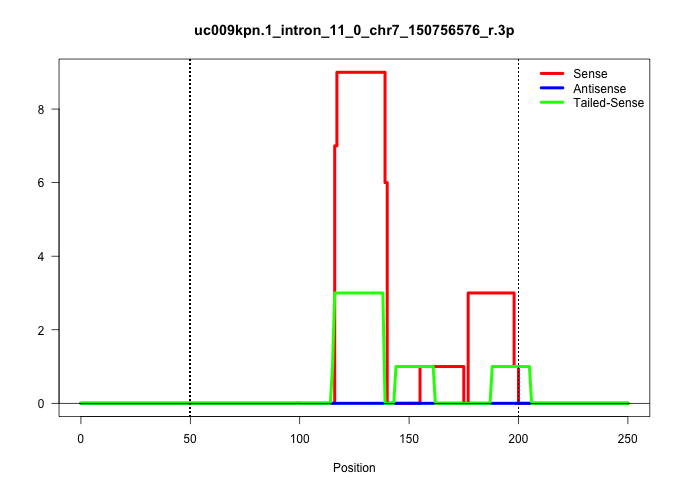
| Gene: Cars | ID: uc009kpn.1_intron_11_0_chr7_150756576_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(11) TESTES |
| GGATCTAGAAATAATTTCGAGACTCTAGCTCAGCCACAGCACAGACTGACTGAACCATATAAACAGGTTTTCCCTTGGTTTATTCCTCTGAAGATTTTGAGCTGGTGCAGTGAGGTTCAGAGGCATAAAGGTCATTTGCTTCAGTAGTAAAGGCCTGGGCAGGTTTGGAATGGTCAGCAGGGCCCTTTATGTCTCTCTAGGGTCGTCCCGGATGGCACATAGAGTGTTCTGCTATGGCAGGCACGCTCCT ......................................................................................................................(((((((((((((...(((((((((.......((((((....))))))....)))..))))))..)))))))))))))...................................................... ...............................................................................................................112.....................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGCT.............................................................................................................. | 24 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGC............................................................................................................... | 23 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAGAGGCATAAAGGTCATTTGCT.............................................................................................................. | 23 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCT.................................................... | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGt............................................................................................................... | 23 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTac............................................................................................................... | 23 | ac | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGTAAAGGCCTGGagcc........................................................................................ | 18 | agcc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCTAG.................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................ATGTCTCTCTAGGGTgtg............................................ | 18 | gtg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGGGCAGGTTTGGAATGGTC........................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTGt............................................................................................................... | 24 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................TAAAGGTCATTTGCT.............................................................................................................. | 15 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | 0.08 |
| GGATCTAGAAATAATTTCGAGACTCTAGCTCAGCCACAGCACAGACTGACTGAACCATATAAACAGGTTTTCCCTTGGTTTATTCCTCTGAAGATTTTGAGCTGGTGCAGTGAGGTTCAGAGGCATAAAGGTCATTTGCTTCAGTAGTAAAGGCCTGGGCAGGTTTGGAATGGTCAGCAGGGCCCTTTATGTCTCTCTAGGGTCGTCCCGGATGGCACATAGAGTGTTCTGCTATGGCAGGCACGCTCCT ......................................................................................................................(((((((((((((...(((((((((.......((((((....))))))....)))..))))))..)))))))))))))...................................................... ...............................................................................................................112.....................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|