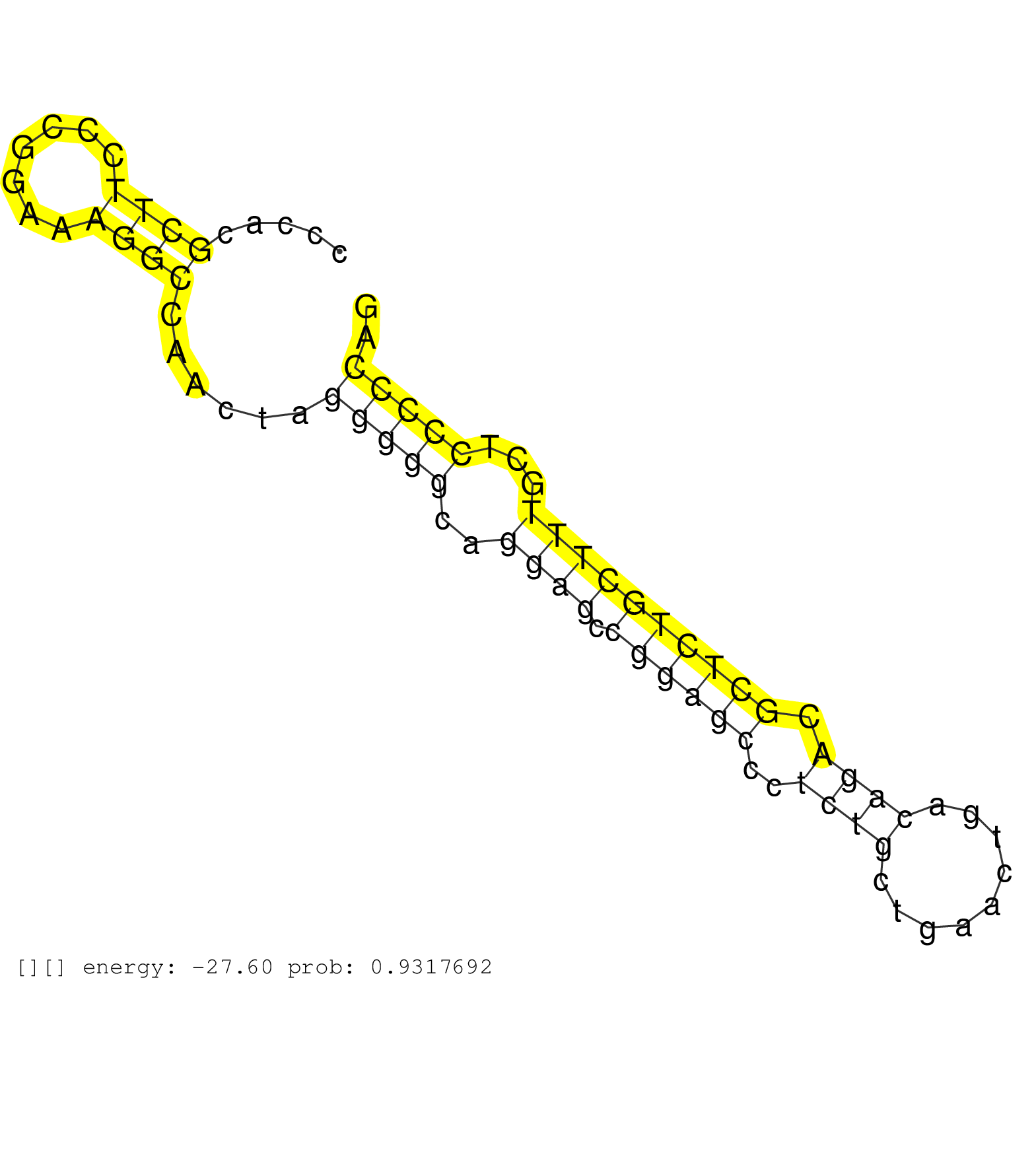
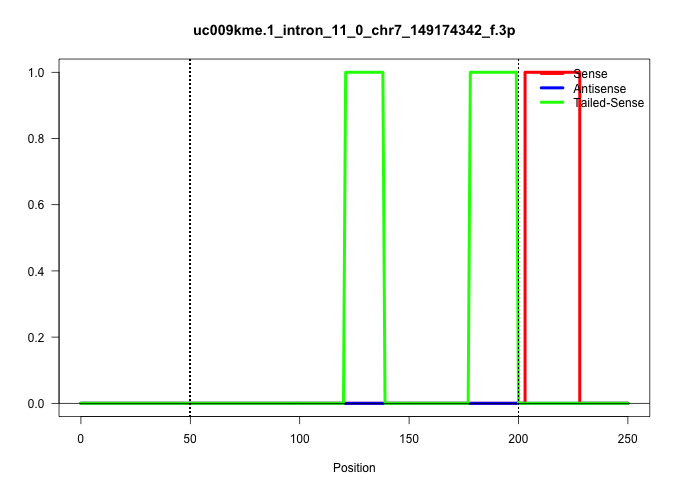
| Gene: Brsk2 | ID: uc009kme.1_intron_11_0_chr7_149174342_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(7) TESTES |
| AGCCCCAGGGACCCTGGATACATACTGGGTGGTGACAGCCAAGGTCAGCAGGATGGCCCCAGGACACATCTGTGCCTCTAGAGACCTTCCTCGGCCTTGGAGCTCCGGGTCAGGACCCCACGCTTCCCGGAAAGGCCAACTAGGGGGCAGGAGCCGGAGCCCTCTGCTGAACTGACAGACGCTCTGCTTTGCTCCCCCAGATCTCGATCCATCAGTGGTGCGTCCTCAGGCCTTTCTACAAGTCCACTCA .........................................................................................................................((((.......))))......(((((..((((.((((((..((((.........)))).))))))))))...))))).................................................... ....................................................................................................................117................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................ACGCTCTGCTTTGCTCCCCCAt.................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................................................................TCGATCCATCAGTGGTGCGTCCTCA...................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................ACGCTCTGCTTTGCTCCCCCAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................GCTTCCCGGAAAGGCtgt............................................................................................................... | 18 | tgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - |
| AGCCCCAGGGACCCTGGATACATACTGGGTGGTGACAGCCAAGGTCAGCAGGATGGCCCCAGGACACATCTGTGCCTCTAGAGACCTTCCTCGGCCTTGGAGCTCCGGGTCAGGACCCCACGCTTCCCGGAAAGGCCAACTAGGGGGCAGGAGCCGGAGCCCTCTGCTGAACTGACAGACGCTCTGCTTTGCTCCCCCAGATCTCGATCCATCAGTGGTGCGTCCTCAGGCCTTTCTACAAGTCCACTCA .........................................................................................................................((((.......))))......(((((..((((.((((((..((((.........)))).))))))))))...))))).................................................... ....................................................................................................................117................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................TCCATCAGTGGTGCGTCCTCAGGCCTTTCTA............ | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................CTTTCTACAAGTCCAcct.... | 18 | cct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................CTTTGCTCCCCCAGATCTCGATCCA....................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................CCATCAGTGGTGCGTCCTCAGGCCTTTCTAt............ | 31 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - |