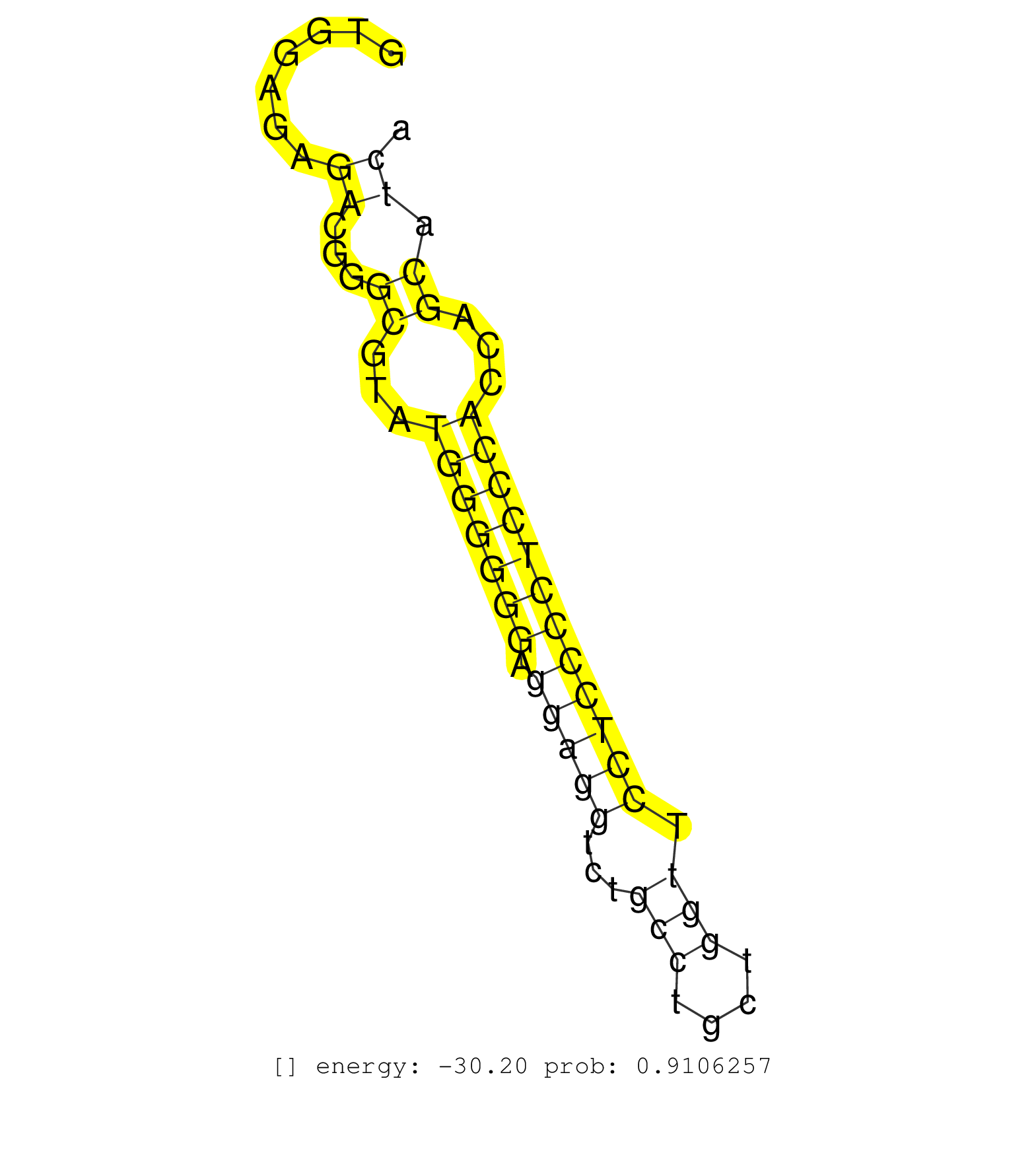

| Gene: Slc25a22 | ID: uc009kkz.1_intron_2_0_chr7_148617556_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(23) TESTES |
| AATCATGGCATTGCTGGCCTCTACAAGGGACTGGGGGCCACGCTACTCAGGTGGAGAGACGGGCGTATGGGGGGAGGAGGTCTGCCTGCTGGTTCCTCCCCTCCCACCAGCATCACTCTTCCCTCCCCACAGGGATGTTCCCTTCTCCATTGTCTACTTCCCCCTGTTTGCCAACCTGAATC .........................................................((...((...(((((((.(((((...(((....))).))))))))))))...)).)).................................................................... ..................................................51..............................................................115................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGAGAGACGGGCGTATGGGGGGA........................................................................................................... | 25 | 1 | 16.00 | 16.00 | 5.00 | 3.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGAG.......................................................................................................... | 26 | 1 | 15.00 | 15.00 | 5.00 | 3.00 | - | - | 2.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGG............................................................................................................ | 24 | 1 | 6.00 | 6.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGG............................................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............GGCCTCTACAAGGGACTGGG................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TACAAGGGACTGGGGGCCACGCTACT....................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGAt.......................................................................................................... | 26 | t | 1.00 | 16.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGGACTGGGGGCCACGCT.......................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCTCCATTGTCTACTTCCCCCTGTT.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................ACAAGGGACTGGGGGCCACGCTACTCAG.................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................TCCTCCCCTCCCACCgcg....................................................................... | 18 | gcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTGTCTACTTCCCCCTGTTTGCCAAC....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGg........................................................................................................... | 25 | g | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGG............................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGa............................................................................................................ | 24 | a | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGAaa......................................................................................................... | 27 | aa | 1.00 | 16.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TACAAGGGACTGGGGGCCACGCTACTCAG.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGc........................................................................................................... | 25 | c | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGAGAGACGGGCGTATGGGGGGAGG......................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGAa.......................................................................................................... | 26 | a | 1.00 | 16.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ACAAGGGACTGGGGGCCACG............................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGGAGAGACGGGCGTATGGGGGGt........................................................................................................... | 25 | t | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................CTCTACAAGGGACTGGGG.................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| AATCATGGCATTGCTGGCCTCTACAAGGGACTGGGGGCCACGCTACTCAGGTGGAGAGACGGGCGTATGGGGGGAGGAGGTCTGCCTGCTGGTTCCTCCCCTCCCACCAGCATCACTCTTCCCTCCCCACAGGGATGTTCCCTTCTCCATTGTCTACTTCCCCCTGTTTGCCAACCTGAATC .........................................................((...((...(((((((.(((((...(((....))).))))))))))))...)).)).................................................................... ..................................................51..............................................................115................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................GCTGGTTCCTCCCCTggg................................................................................ | 18 | ggg | 3.00 | 0.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GCTGGTTCCTCCCCTCCgggc.............................................................................. | 21 | gggc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TCCCTTCTCCATTGTCTACTTCCCCCT................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GCTGGTTCCTCCCCTCCggg.............................................................................. | 20 | ggg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCCCACAGGGATGTTCCCTTCTCCATTGTCTA.......................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................CCACAGGGATGTTCCCT....................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| ...................................................................................GCCTGCTGGTTCCTCC................................................................................... | 16 | 9 | 0.11 | 0.11 | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |