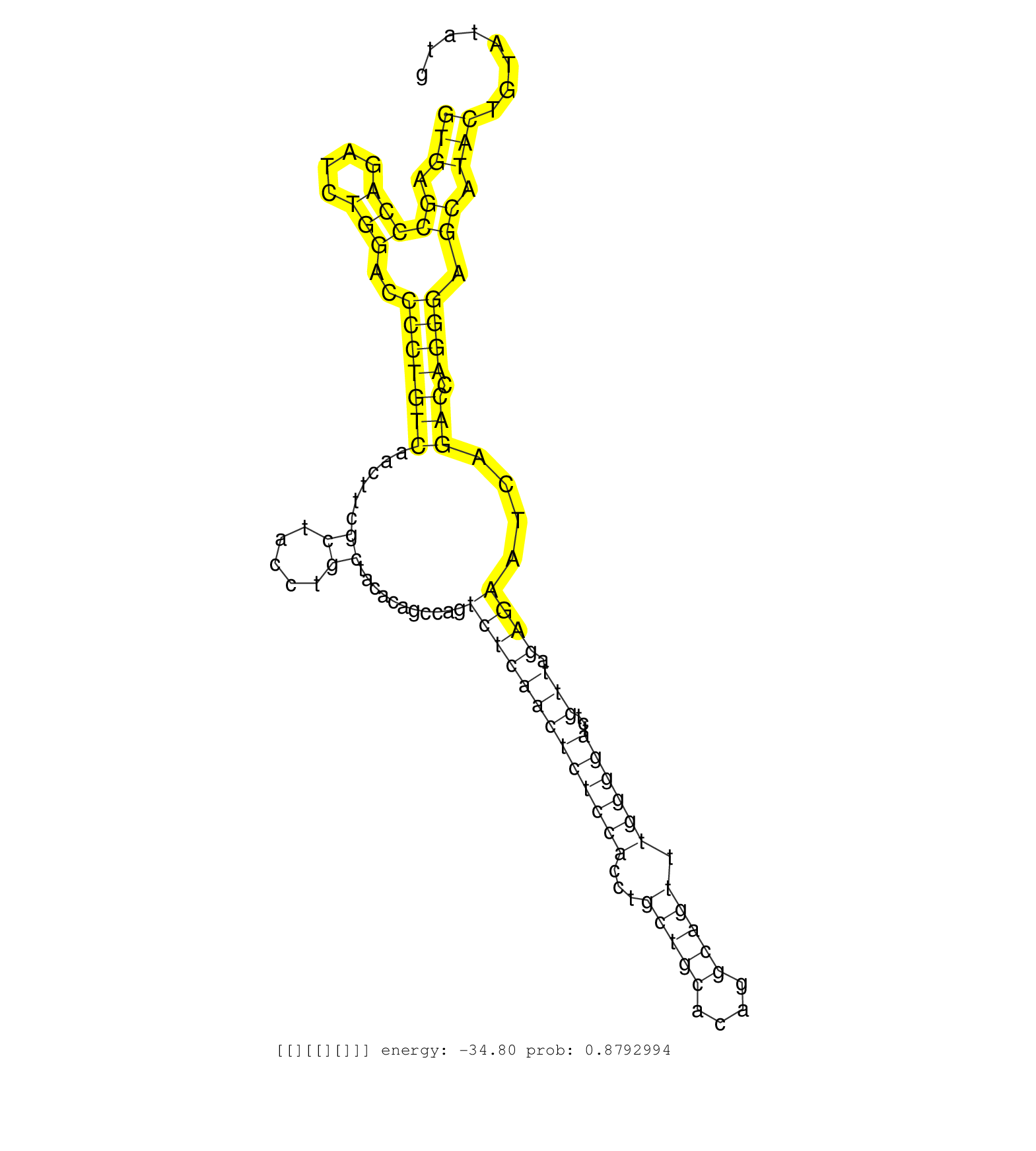

| Gene: Rassf7 | ID: uc009kka.1_intron_2_0_chr7_148403612_f | SPECIES: mm9 |
 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| CCTTGGCCTTGGTGAGCCAGGCTCTGGAGGCTGCTGAGCACGCTCTGCAGGTGAGCCCAGATCTGGACCCCTGTCAACTTCGCTACCTGCTACACAGCCAGTCTCAACTCTCCACCTGCTGCACAGGCAGTTTGGGGATCTGTTAGAGAATCAGACCAGGGAGCATACTGTATATGGAAAAGAAGGTTGTGGCCTCGTCCCTGAAGGTCCTCTGTACCCCCAGGCCCAGGCTCAGGAGCTGGAGGAGCTGAACAGGGAACTCCGTCAGTGCAA ..................................................(((.(((((....)))..(((((((......((.....))...........(((((((((((((...(((((....))))).))))))...))).))))....))).)))).)).)))......................................................................................................... ..................................................51...........................................................................................................................176............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................AGAATCAGACCAGGGAGCATACTGTA..................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........TGGTGAGCCAGGCTCTGGAGGCTGC............................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TAGAGAATCAGACCAGGGAGCATACT........................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTTAGAGAATCAGACCAGGGAGCAT........................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................ACCAGGGAGCATACTGTATATGGAAAAGt.......................................................................................... | 29 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................TGAACAGGGAACTCCGTCAGTGCAA | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................AGAAGGTTGTGGCCTCGTCCCTGAAG................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TACTGTATATGGAAAAGAAGGTTGTGGC................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TAGAGAATCAGACCAGGGAGCATAC......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TACTGTATATGGAAAAGAAGGTTGTG.................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TAGAGAATCAGACCAGGGAGCATACTGaa..................................................................................................... | 29 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TCAGGAGCTGGAGGAGCTGAAC.................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GGCAGTTTGGGGATCTGTTAGAGAATC......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATCTGTTAGAGAATCAGACCAGGGAG.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTAGAGAATCAGACCAGGGAGCATACTGT...................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GAATCAGACCAGGGAGCATACTGTATA................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TATATGGAAAAGAAGGTTGTGGCCTCGTC.......................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TCAGGAGCTGGAGGAGCTGAACAGG................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................TGAACAGGGAACTCCGTCAGTGtttt | 26 | tttt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TAGAGAATCAGACCAGGGAGCATACTG....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................TGGAGGCTGCTGAGCACGCTCTGC................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................AATCAGACCAGGGAGCATACTGTAT.................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................TACTGTATATGGAAAAGAAGGTTGTGG................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CTGTTAGAGAATCAGACCAGGGAGCA............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................AATCAGACCAGGGAGCATACTGTATA................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCCAGATCTGGACCCCTGTC...................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................ATCAGACCAGGGAGCATACTGTATAT.................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................GGCTCAGGAGCTGGAGGAGCTGA...................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| CCTTGGCCTTGGTGAGCCAGGCTCTGGAGGCTGCTGAGCACGCTCTGCAGGTGAGCCCAGATCTGGACCCCTGTCAACTTCGCTACCTGCTACACAGCCAGTCTCAACTCTCCACCTGCTGCACAGGCAGTTTGGGGATCTGTTAGAGAATCAGACCAGGGAGCATACTGTATATGGAAAAGAAGGTTGTGGCCTCGTCCCTGAAGGTCCTCTGTACCCCCAGGCCCAGGCTCAGGAGCTGGAGGAGCTGAACAGGGAACTCCGTCAGTGCAA ..................................................(((.(((((....)))..(((((((......((.....))...........(((((((((((((...(((((....))))).))))))...))).))))....))).)))).)).)))......................................................................................................... ..................................................51...........................................................................................................................176............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................CTGTTAGAGAATCAgatc........................................................................................................................ | 18 | gatc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................GCTACCTGCTACACA................................................................................................................................................................................. | 15 | 11 | 0.73 | 0.73 | - | - | - | 0.09 | 0.36 | - | - | - | - | - | - | - | 0.27 |