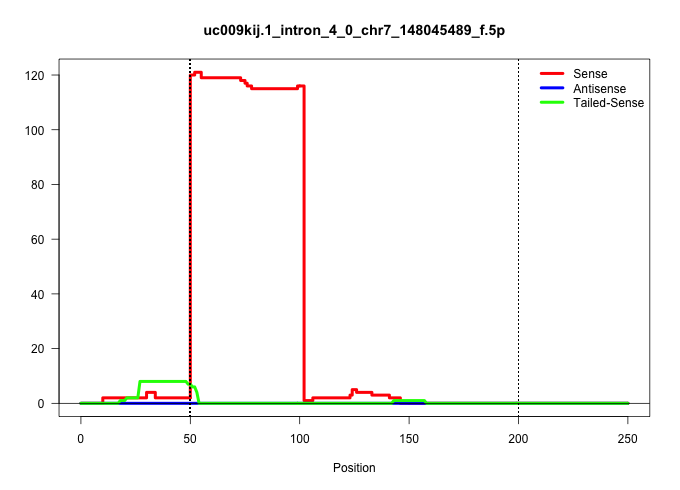
| Gene: Ric8 | ID: uc009kij.1_intron_4_0_chr7_148045489_f.5p | SPECIES: mm9 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(12) TESTES |
| TGGATGTGATCAGTGCCCTCCTCGCCTTCCTAGAGAAACGTCTGCACCAGGTAGGACTGGGGGCCTGCTACGTGGACACCCAGGGGCTCGGGCACCTGTTGTGACTGCTGTTTCTGAACCATTGGTACTGGCGAGTAGAACTCAGCTTGACGATATATCGTGGACAGTTGCTCTGACTGCTCTGTGGTGCAGCTAGACGGTGGCCGCTCAGTCAGGCCCTCGCAGAAGCCTGCAGATACACTGTGTGTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGACTGGGGGCCTGCTACGTGGACACCCAGGGGCTCGGGCACCTGTTGT.................................................................................................................................................... | 52 | 1 | 115.00 | 115.00 | 115.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCTAGAGAAACGTCTGCACCAGaccc.................................................................................................................................................................................................... | 27 | accc | 4.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - |
| ............................................................................................................................GTACTGGCGAGTAGAACTCAGC........................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCTAGAGAAACGTCTGCACCAGacc..................................................................................................................................................................................................... | 26 | acc | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ..........CAGTGCCCTCCTCGCCTTCCTAGA........................................................................................................................................................................................................................ | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TAGAGAAACGTCTGCACCAGGTAGG................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAGGACTGGGGGCCTGCTACGT................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................TCGCCTTCCTAGAGAAACGTCTGCACCAGa....................................................................................................................................................................................................... | 30 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................AGGACTGGGGGCCTGCTACGTGGACA............................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................GGTACTGGCGAGTAGAAC............................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGTGACTGCTGTTTCTGAACCATTGGT............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................TCCTCGCCTTCCTAGAGAAACGTCTGCACCt......................................................................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GCTGTTTCTGAACCATTGGTACTGGCG..................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAGGACTGGGGGCCTGCTACGTGG............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGGGGGCCTGCTACGTGGA.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AGCTTGACGATtcga............................................................................................ | 15 | tcga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TGGATGTGATCAGTGCCCTCCTCGCCTTCCTAGAGAAACGTCTGCACCAGGTAGGACTGGGGGCCTGCTACGTGGACACCCAGGGGCTCGGGCACCTGTTGTGACTGCTGTTTCTGAACCATTGGTACTGGCGAGTAGAACTCAGCTTGACGATATATCGTGGACAGTTGCTCTGACTGCTCTGTGGTGCAGCTAGACGGTGGCCGCTCAGTCAGGCCCTCGCAGAAGCCTGCAGATACACTGTGTGTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................GCTCAGTCAGGCCCTttga.............................. | 19 | ttga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |