
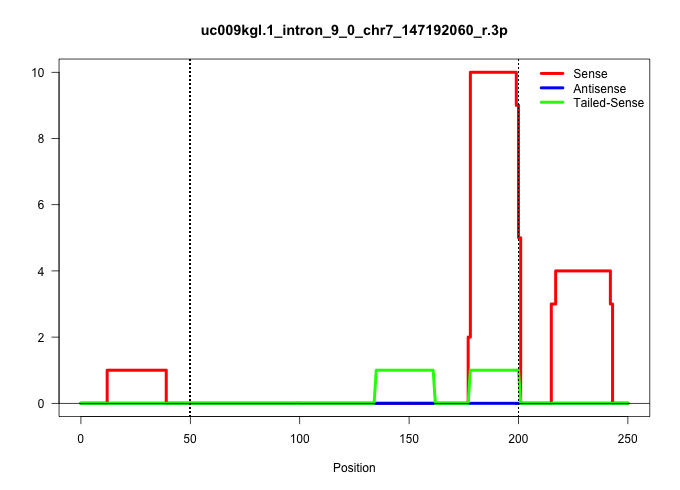
| Gene: Tubgcp2 | ID: uc009kgl.1_intron_9_0_chr7_147192060_r.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| GTGTCTCTCCTGTGGCCATAATGGAAGAGTAGGGAGGAAGCTACTCAGCTGGGTCTCAAATGAGACACCTGCTCCCAAGTAAGGCTCAGTTGCTCAGCTCACAGTTGAGTGGTTGTGCAGGACGGTCCTGGAAGTATGTGGCCACACCAGAGTTGTCTGGGCTTGTGCTGTAGTGGCCTGAAACTCTGTGTGACTTACAGTGAGTTCATGGTGGAGGAACACGAGCTGCGGAAGGAGAAGATCCAGGAGG ........................................................................................................................................(((((.((((.(((((((.((.(((((...((....)))))))))))))))))))).)).)))................................................... ..................................................................................................................................131..................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TGAAACTCTGTGTGACTTACAGT................................................. | 23 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................TGAAACTCTGTGTGACTTACAG.................................................. | 22 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................GGAACACGAGCTGCGGAAGGAGAAGATC....... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................ATGTGGCCACACCAGAGTTGTCTGGat........................................................................................ | 27 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................CTGAAACTCTGTGTGACTTACA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AACACGAGCTGCGGAAGGAGAAGATC....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGAGTTCATGGTGGAGGAACACGAGCTGCGGAAG................ | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGAAACTCTGTGTGACTTAC.................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............TGGCCATAATGGAAGAGTAGGGAGGAA................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................CTGAAACTCTGTGTGACTTACAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GGAACACGAGCTGCGGAAGGAGAAGAT........ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TACAGTGAGTTCATGGTGGAGGAACACG........................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................TGAAACTCTGTGTGACTTACA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GAACACGAGCTGCGGAAGGAGAAGA......... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGAAACTCTGTGTGACTTACAGa................................................. | 23 | a | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTGTCTCTCCTGTGGCCATAATGGAAGAGTAGGGAGGAAGCTACTCAGCTGGGTCTCAAATGAGACACCTGCTCCCAAGTAAGGCTCAGTTGCTCAGCTCACAGTTGAGTGGTTGTGCAGGACGGTCCTGGAAGTATGTGGCCACACCAGAGTTGTCTGGGCTTGTGCTGTAGTGGCCTGAAACTCTGTGTGACTTACAGTGAGTTCATGGTGGAGGAACACGAGCTGCGGAAGGAGAAGATCCAGGAGG ........................................................................................................................................(((((.((((.(((((((.((.(((((...((....)))))))))))))))))))).)).)))................................................... ..................................................................................................................................131..................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................CCTGGAAGTATGTGGCCAcaga.......................................................................................................... | 22 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................GCTACTCAGCTGGGTgaaa.................................................................................................................................................................................................... | 19 | gaaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |