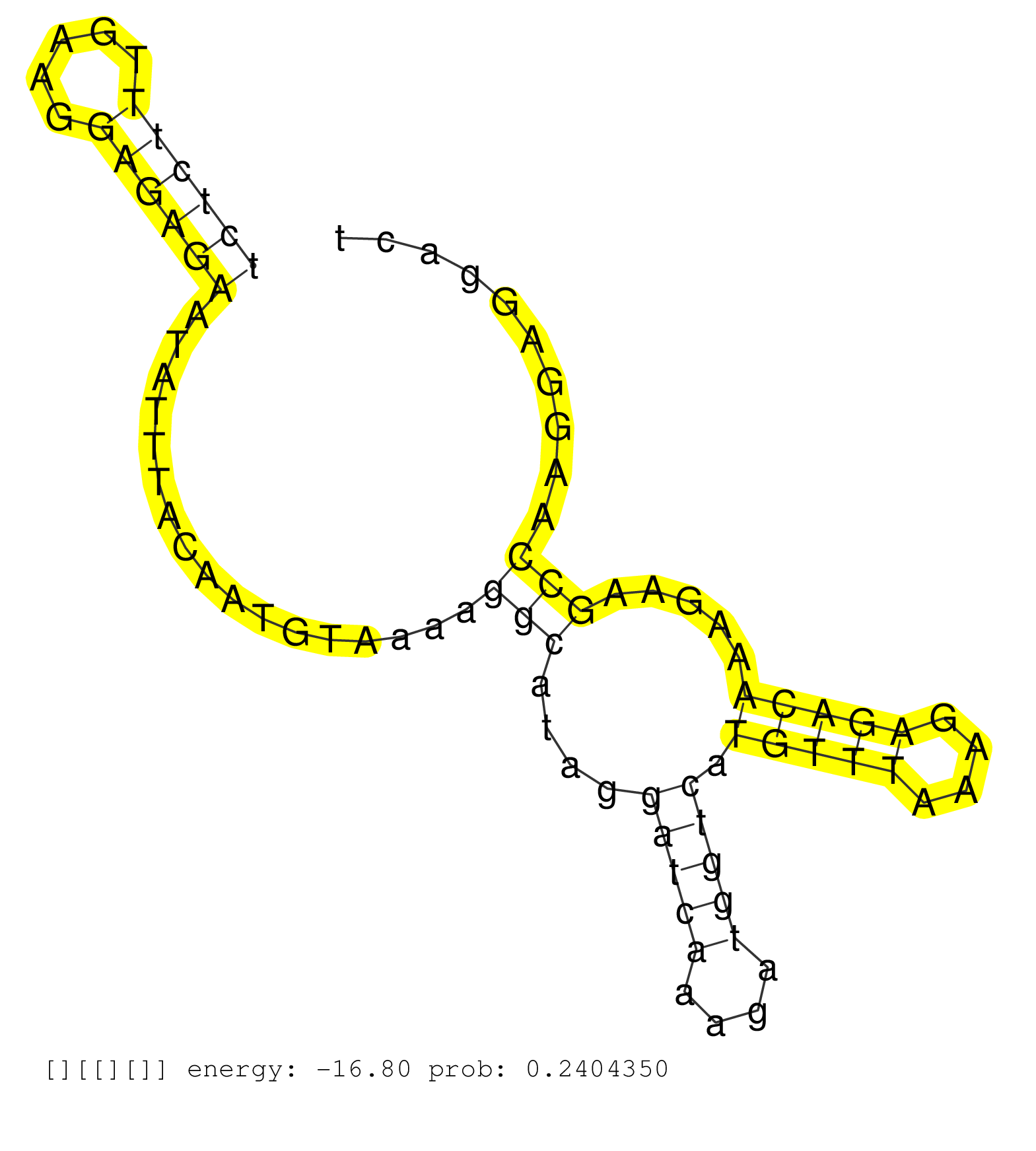
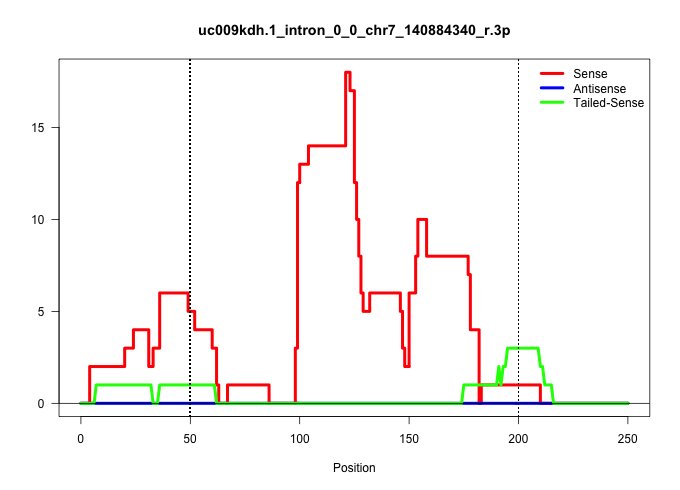
| Gene: Uros | ID: uc009kdh.1_intron_0_0_chr7_140884340_r.3p | SPECIES: mm9 |
 |
 |
|
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(15) TESTES |
| TATTTACTATGAGACAGGCATCGCTGTGGGCACTTATAGCTGTTGACTCGTTTAACCTGGTCACTCATTATAGACAGCGTCCTCTCACTGGCCCTCTCTTTGAAGGAGAGAATATTTACAATGTAAAAGGCATAGGATCAAAGATGGTCATGTTTAAAGAGACAAAGAAGCCAAGGAGGACTGTACTACCTTTGTTTCAGATGGCCTCCTGGGTGAACAGCGATCCCGCTCCCATGCTCCAGGTAGCCTT ..............................................................................................((((((.....)))))).................(((....(((((....))))).(((((....))))).....))).............................................................................. ..............................................................................................95.....................................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................TGTTTAAAGAGACAAAGAAGCCAAGGAG........................................................................ | 28 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTGAAGGAGAGAATATTTACAATGTA............................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....TACTATGAGACAGGCATCGCTGTGGGC........................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TAAAGAGACAAAGAAGCCAAGGAGGACT.................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGTAAAAGGCATAGGATCAAAGATGG....................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTGAAGGAGAGAATATTTACAATGTAAAA.......................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TAGCTGTTGACTCGTTTAACCTGGTC............................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................TTTGAAGGAGAGAATATTTACAATGTA............................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TTAAAGAGACAAAGAAGCCAAGGAGGACT.................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TAGGATCAAAGATGGTCATGTTTAAA............................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTGAAGGAGAGAATATTTACAATGTAA............................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................TTCAGATGGCCTCCcaa...................................... | 17 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................GTTTCAGATGGCCTatt........................................ | 17 | att | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TAGCTGTTGACTCGTTTAACCTGGTCA........................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTATAGACAGCGTCCTCTC.................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GAGGACTGTACTAtcat.......................................................... | 17 | tcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTGTTTCAGATGGCCTCCTGGGTGt.................................. | 25 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................TGTGGGCACTTATAGCTGTTGACTCGTT...................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TAGCTGTTGACTCGTTTAACCTGGTa............................................................................................................................................................................................ | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................TTGAAGGAGAGAATATTTACAATGTAAA........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......TATGAGACAGGCATCGCTGTGGGCta......................................................................................................................................................................................................................... | 26 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................TTGAAGGAGAGAATATTTACAATGTAAAAG......................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................TGTAAAAGGCATAGGATCAAAGATGGT...................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TACTACCTTTGTTTCAGATGGCCTCCT........................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TGAAGGAGAGAATATTTACAATGTAAA........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGTAAAAGGCATAGGATCAAAGATG........................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................TTATAGCTGTTGACTCGTTTAACCTGG.............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TGTTTAAAGAGACAAAGAAGCCAAGGA......................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTTGAAGGAGAGAATATTTACAATG............................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................GGAGAGAATATTTACAATGTAAAAGGCA...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TCGCTGTGGGCACTTATAGCTGTTGACTC......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TATTTACTATGAGACAGGCATCGCTGTGGGCACTTATAGCTGTTGACTCGTTTAACCTGGTCACTCATTATAGACAGCGTCCTCTCACTGGCCCTCTCTTTGAAGGAGAGAATATTTACAATGTAAAAGGCATAGGATCAAAGATGGTCATGTTTAAAGAGACAAAGAAGCCAAGGAGGACTGTACTACCTTTGTTTCAGATGGCCTCCTGGGTGAACAGCGATCCCGCTCCCATGCTCCAGGTAGCCTT ..............................................................................................((((((.....)))))).................(((....(((((....))))).(((((....))))).....))).............................................................................. ..............................................................................................95.....................................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|