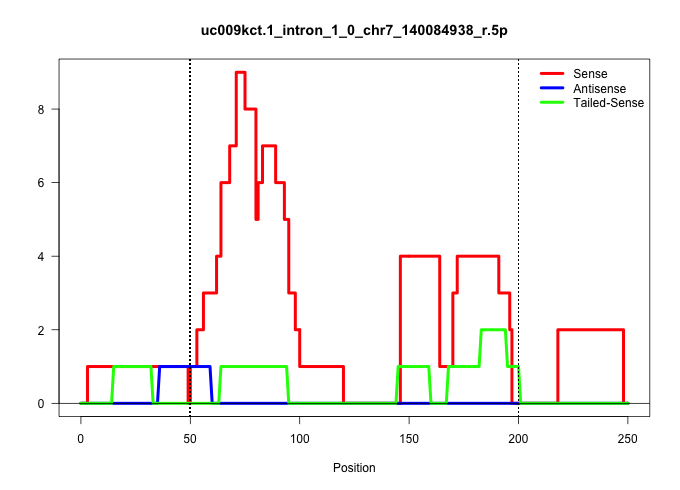
| Gene: AK005115 | ID: uc009kct.1_intron_1_0_chr7_140084938_r.5p | SPECIES: mm9 |
 |
|
(7) OTHER.mut |
(1) PIWI.ip |
(16) TESTES |
| TCCTGATTGTGAGGCTACGGCGCGCGCGGTCCCGCGACGCGTCCTCCCGGGTGAGTGTCTTCAAGTCTAAGTGCAGGATGCACTTGGGAGGTGGGAGCCTTCTTCCCCTGGCCGCCGGCCGCTCTTTGCTGCAGAGTCGCCAGCGCTTTGAATCAAGCGTGAGTCACGCATCTGTTTGCTAACTGGGAGACCGCTTGGCCGTTCCTTCGGCGGTTTAAAACAGTTTTTAAAATTTCAATAATTTGACCCA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TTTGAATCAAGCGTGAGT...................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................AACAGTTTTTAAAATTTCAATAATTTGACC.. | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GTCTAAGTGCAGGATGCACTTGGGAGGTGGG........................................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................ACTTGGGAGGTGGGAGCCT...................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................TTGGGAGGTGGGAGCCTT..................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTCTTCAAGTCTAAGTGCAGGATG.......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................TCTGTTTGCTAACTGGGAGACCGCTTG..................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............TACGGCGCGCGCGGgccc......................................................................................................................................................................................................................... | 18 | gccc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................CCCGCGACGCGTCCTCCCG......................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................GTCTTCAAGTCTAAGTGCA............................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................TCTGTTTGCTAACTGGGAGACCGCTT...................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAGTCTAAGTGCAGGATG.......................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GTCTAAGTGCAGGATGCACTTGGGAGGgggg........................................................................................................................................................... | 31 | gggg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGTTTGCTAACTGGGAGACCGCTTG..................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................AAGTGCAGGATGCACTTGGGAGGTGGGAGC........................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGATTGTGAGGCTACGGCGCGCGCGGT............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................TGCAGGATGCACTTGGGA................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CATCTGTTTGCTAACTGGGAGACCGCa....................................................... | 27 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CTTCCCCTGGCCGCCGGCC.................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGGGAGACCGCTTGGgaa................................................. | 18 | gaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGTGTCTTCAAGTCTAAGTGCAGGATG.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................GTTTGCTAACTGGGAGAC........................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTTGAATCAAGCGTGAGTCACGCATCT............................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CTTTGAATCAAGCGa.......................................................................................... | 15 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGCAGGATGCACTTGGGAGGTG............................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCTGATTGTGAGGCTACGGCGCGCGCGGTCCCGCGACGCGTCCTCCCGGGTGAGTGTCTTCAAGTCTAAGTGCAGGATGCACTTGGGAGGTGGGAGCCTTCTTCCCCTGGCCGCCGGCCGCTCTTTGCTGCAGAGTCGCCAGCGCTTTGAATCAAGCGTGAGTCACGCATCTGTTTGCTAACTGGGAGACCGCTTGGCCGTTCCTTCGGCGGTTTAAAACAGTTTTTAAAATTTCAATAATTTGACCCA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................ACGCGTCCTCCCGGGTGAGTGTCT.............................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |