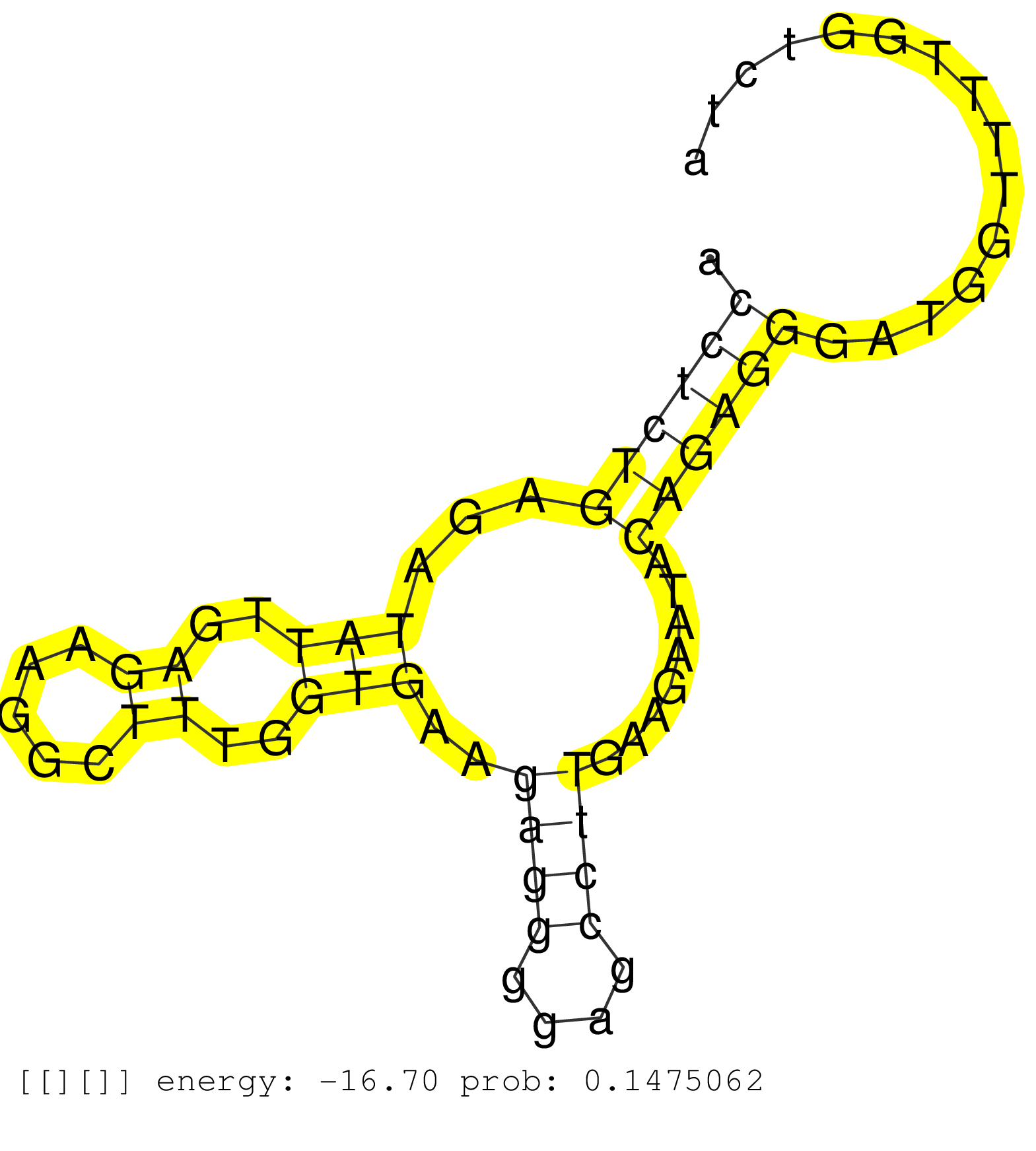
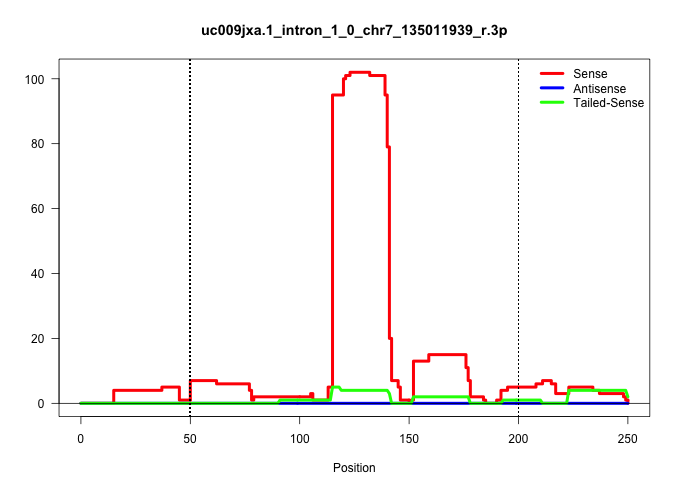
| Gene: Zfp668 | ID: uc009jxa.1_intron_1_0_chr7_135011939_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| TTCCCAGCACCGACATGGTAGCTCACAGCCTTAGGCACTAAGCATAGGCATGGGCCAGTAAACATTCATACATGGAAAATGAAAGTGATATTTTCCTTAAAGGTAGGGTCACCTCTGAGATATTGAGAAGGCTTTGGTGAAGAGGGGAGCCTTGAAGAATACAGAGGGATGGTTTTGGTCTAATTTTGCTTCTTTTGCAGAGAAGGCCTACTCACCAAGGCCATGGAAGTGGAGGCTACAGAGGCCAGGT ...............................................................................................................((((((...(((..((.....))..)))..((((....))))........))))))................................................................................... ..............................................................................................................111....................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTGAA............................................................................................................. | 26 | 1 | 59.00 | 59.00 | 16.00 | 20.00 | 4.00 | 7.00 | 4.00 | 3.00 | 1.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTGA.............................................................................................................. | 25 | 1 | 16.00 | 16.00 | 5.00 | 3.00 | 3.00 | - | 1.00 | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTGAAG............................................................................................................ | 27 | 1 | 13.00 | 13.00 | 3.00 | - | 2.00 | - | 1.00 | - | 1.00 | 3.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................TGAAGAATACAGAGGGATGGTTTTGG........................................................................ | 26 | 1 | 5.00 | 5.00 | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...............TGGTAGCTCACAGCCTTAGGCACTAAGCAT............................................................................................................................................................................................................. | 30 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCTGAGATATTGAGAAGGCTTTGGTG............................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGAAGAATACAGAGGGATGGTTTTG......................................................................... | 25 | 1 | 4.00 | 4.00 | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TATTGAGAAGGCTTTGGTGAAGAGGG........................................................................................................ | 26 | 1 | 4.00 | 4.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGAAGAATACAGAGGGATGGTTTT.......................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TGGGCCAGTAAACATTCATACATGGAAA............................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTTTGCAGAGAAGGCCTACTCACCA................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGAAGAATACAGAGGGATGGTTTTGt........................................................................ | 26 | t | 2.00 | 4.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTGAAa............................................................................................................ | 27 | a | 2.00 | 59.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TGGGCCAGTAAACATTCATACATGGAA............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTG............................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGAAAGTGATATTTTCCTTAAAGGTAG................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGAAGTGGAGGCTACAGAGGCCAGGTa | 28 | a | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TATTGAGAAGGCTTTGGTGAAGAGG......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TACAGAGGGATGGTTTTGGTCTAATT................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CTAAGCATAGGCATGGGCCAGTAAA............................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ATTGAGAAGGCTTTGGTGAAGAGG......................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TGGGCCAGTAAACATTCATACATGGAAAA........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTGAt............................................................................................................. | 26 | t | 1.00 | 16.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAGATATTGAGAAGGCTTTGGTGAAt............................................................................................................ | 27 | t | 1.00 | 59.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTTCCTTAAAGGTAGGGTCACCTCTGAa................................................................................................................................... | 28 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGAAGTGGAGGCTACAGAGGCCAGG. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTTTGCAGAGAAGGCCTACTCAC................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................................TGGAAGTGGAGGCTACAGAGGCCAG.. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGAGAAGGCCTACTCACCAAGGCCA........................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TACTCACCAAGGCCATGGAAGTGGAG................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................TCACCAAGGCCATGGAAGTGGAGGCT............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGAAGTGGAGGCTACAGAGGCCAGGa | 27 | a | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TTGAAGAATACAGAGGGATGGTTTT.......................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TTTGCAGAGAAGGCgcac....................................... | 18 | gcac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................TGAGAAGGCTTTGGTGAAGAGGGGAGC.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGAAGTGGAGGCTACAGAGGCCAGGg | 27 | g | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TACAGAGGGATGGTTTTGGTCTAAT.................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCTTTTGCAGAGAAGGCCTACTCACCA................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGAAGTGGAGGCTACAGAGGCCAGGT | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GGGTCACCTCTGAGATATTGAGAAGGC...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTCCCAGCACCGACATGGTAGCTCACAGCCTTAGGCACTAAGCATAGGCATGGGCCAGTAAACATTCATACATGGAAAATGAAAGTGATATTTTCCTTAAAGGTAGGGTCACCTCTGAGATATTGAGAAGGCTTTGGTGAAGAGGGGAGCCTTGAAGAATACAGAGGGATGGTTTTGGTCTAATTTTGCTTCTTTTGCAGAGAAGGCCTACTCACCAAGGCCATGGAAGTGGAGGCTACAGAGGCCAGGT ...............................................................................................................((((((...(((..((.....))..)))..((((....))))........))))))................................................................................... ..............................................................................................................111....................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|