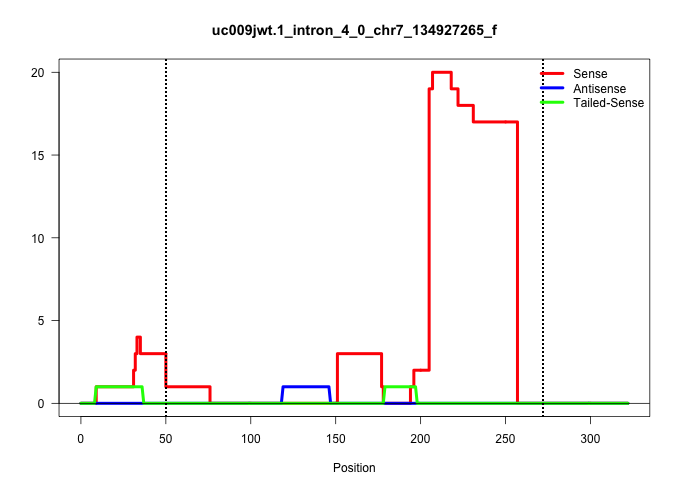
| Gene: Setd1a | ID: uc009jwt.1_intron_4_0_chr7_134927265_f | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| GCAAGGCCTTGAGTGAGAAGTTCCAGGGTTCTGGTGCAGCCGCTGAGACGGTGAGAAGCTTGTAGCTGCCGCTGTGTTAGGCACATGTACTCGTTACCTGGTCACCAAAGCAAGAGCCATTTGGGAAGGCCCTGAAGTCCCCTATTAAGTATAGGAAGAACTAAGGGAAGATGTTTAGGTGTCTTCAGATACAAAGCTGCAATCTCGGATTGGGGCAGTAGACAGTTTTAAATCGTGCCTGATCTTTTTTCCTTGACTCCCTTTTTCTTTAGACTGAAGCCCGCCGACGGTCTTCCTCGGACACAGCTGCCTATCCAGCAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................CGGATTGGGGCAGTAGACAGTTTTAAATCGTGCCTGATCTTTTTTCCTTGAC................................................................. | 52 | 1 | 17.00 | 17.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAGGAAGAACTAAGGGAAGATGTTTA................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................TGTCTTCAGATACAAAaaa............................................................................................................................ | 19 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAGGAAGAACTAAGGGAAGATGTTTAG................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GATTGGGGCAGTAGACAGTTTTAA........................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................GTGCAGCCGCTGAGACG................................................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CTGCAATCTCGGATTGGGGCAGTAGA.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................AGCTGCAATCTCGGATTGGGGCAG........................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........TGAGTGAGAAGTTCCAGGGTTCTGGT............................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAGTGAGAAGTTCCAGGGTTCTGGTGt............................................................................................................................................................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGAGAAGCTTGTAGCTGCCGCTGTG...................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGGTGCAGCCGCTGAGACG................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GGTGCAGCCGCTGAGACG................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCAAGGCCTTGAGTGAGAAGTTCCAGGGTTCTGGTGCAGCCGCTGAGACGGTGAGAAGCTTGTAGCTGCCGCTGTGTTAGGCACATGTACTCGTTACCTGGTCACCAAAGCAAGAGCCATTTGGGAAGGCCCTGAAGTCCCCTATTAAGTATAGGAAGAACTAAGGGAAGATGTTTAGGTGTCTTCAGATACAAAGCTGCAATCTCGGATTGGGGCAGTAGACAGTTTTAAATCGTGCCTGATCTTTTTTCCTTGACTCCCTTTTTCTTTAGACTGAAGCCCGCCGACGGTCTTCCTCGGACACAGCTGCCTATCCAGCAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................GGTGTCTTCAGATACAaggg................................................................................................................................. | 20 | aggg | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - |
| .......................................................................................................................TTTGGGAAGGCCCTGAAGTCCCCTATTA............................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |