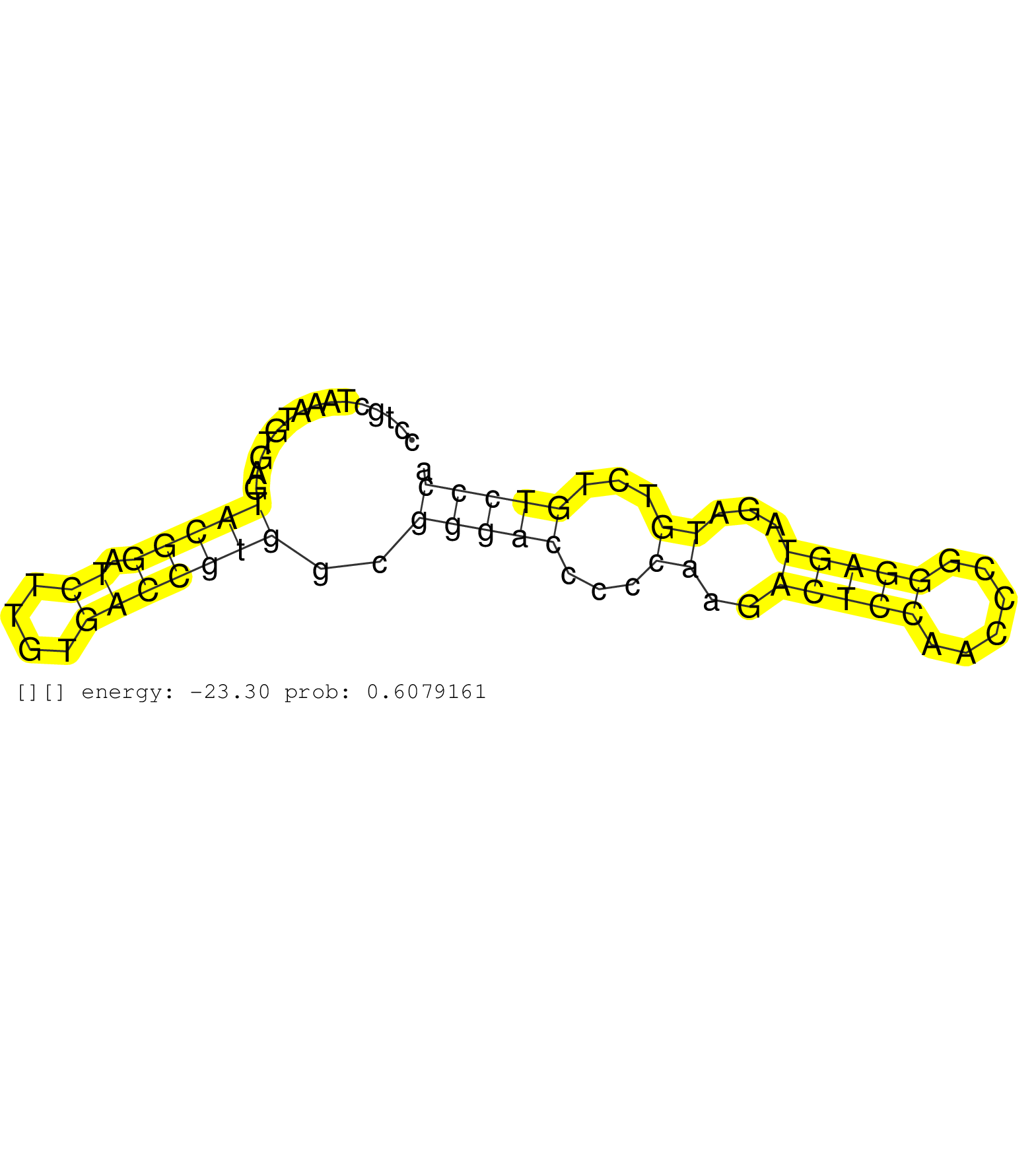

| Gene: Mvp | ID: uc009jtw.1_intron_13_0_chr7_134145473_r.5p | SPECIES: mm9 |
 |
 |
|
(5) PIWI.ip |
(3) PIWI.mut |
(21) TESTES |
| GAGGGAAATTCCTGACCTGTGGCCTGCAGCAGCCAGCTACCCTGCTAAATGTGAGTACGGATCTTGTGACCGTGGCGGGACCCCCAAGACTCCAACCCGGGAGTAGATGTCTGTCCCACAGGCTCTCTCTAGTCTGACTGCTTTCCTGCGACGTTTCTTCTGGGGCAGTGACGTCTCAGCAGTTACAGGAAGGAGGGCGTGGGCAGCTCTGCCTTGTGAGGTTTCACTTTCTAAGACTTTCAGGGTTCGA .......................................................(((((.((....)))))))..(((((...((..(((((......)))))...))...)))))..................................................................................................................................... ........................................41...........................................................................118.................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................TAAATGTGAGTACGGATCTTGTGACC................................................................................................................................................................................... | 26 | 1 | 19.00 | 19.00 | 1.00 | - | 5.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | 3.00 | - | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - |
| .......................................................................................GACTCCAACCCGGGAGTAGATGTCTGT........................................................................................................................................ | 27 | 1 | 14.00 | 14.00 | 6.00 | 2.00 | - | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGCGACGTTTCTTCTGGGGCAGTGAC.............................................................................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGACCTGTGGCCTGCAGCAGCCAGCTA................................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGCGACGTTTCTTCTGGGGCAGTGACG............................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TAGTCTGACTGCTTTCCTGCGACGTTT.............................................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGAGGTTTCACTTTCTAAGACTTTCA........ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGCTAAATGTGAGTACGGATCTTGTG...................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................TCAGCAGTTACAGGAAGGAGGGCGTG................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TAAATGTGAGTACGGATCTTGTGAC.................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGACCTGTGGCCTGCAGCAGCCAGCT.................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTGAGTACGGATCTTGTGACCGTGGC.............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GACTCCAACCCGGGAGTAGATGTCTGTC....................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................CTAAATGTGAGTACGGATCTTGTGACC................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .........TCCTGACCTGTGGCCTGCAGCAGCCAGC..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGTGACCGTGGCGGGACCCCCAAGAC................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TAAATGTGAGTACGGATCTTGTtacc................................................................................................................................................................................... | 26 | tacc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................AGTTACAGGAAGGAGGGCGTGGat.............................................. | 24 | at | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CCTGACCTGTGGCCTGCAGCAGCCAGCTA................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCCTGACCTGTGGCCTGCAGCAGCCAGCT.................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CAGCAGTTACAGGAAGGAGGGCGTG................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TCTCTCTAGTCTGACTGCTTTCCTGC..................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TTCCTGACCTGTGGCCTGCAGCAGCCAGC..................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CCGGGAGTAGATGTCTGTCCCACAGGCTC............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGCTAAATGTGAGTACGGATCTTGTGA..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................CCCTGCTAAATGTGAGTA................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........TTCCTGACCTGTGGCCTGC............................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATCTTGTGACCGTGGCGGa........................................................................................................................................................................... | 19 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCCTGCGACGTTTCTTCTGGGGCA................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCCTGCGACGTTTCTTCTGGGGCAG.................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................ACGTCTCAGCAGTTACAGGAAGGAGG...................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................AGGAGGGCGTGGGCAGCTCTGCCa.................................... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTACGGATCTTGTGACCGTGGC.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCTAGTCTGACTGCTTTCCTGCGACGT................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CTCTCTAGTCTGACTGCTTTCCTGCG.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGCTAAATGTGAGTACGGATCTTGTGAC.................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGCTAAATGTGAGTACGGATCTTGT....................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................GACTCCAACCCGGGAGTAGATGTCTG......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGAGGTTTCACTTTCTAAGACTTTC......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TCTTGTGACCGTGGCGGGACCCCCAA................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TAAATGTGAGTACGGATCTT......................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................ACTCCAACCCGGGAGTAGATGTCTGTC....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TCTCAGCAGTTACAGGAAGGAGGGCGT.................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................AATGTGAGTACGGATCTTGTGACC................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTTCCTGCGACGTTTCTTCTGGGGCAGT................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| GAGGGAAATTCCTGACCTGTGGCCTGCAGCAGCCAGCTACCCTGCTAAATGTGAGTACGGATCTTGTGACCGTGGCGGGACCCCCAAGACTCCAACCCGGGAGTAGATGTCTGTCCCACAGGCTCTCTCTAGTCTGACTGCTTTCCTGCGACGTTTCTTCTGGGGCAGTGACGTCTCAGCAGTTACAGGAAGGAGGGCGTGGGCAGCTCTGCCTTGTGAGGTTTCACTTTCTAAGACTTTCAGGGTTCGA .......................................................(((((.((....)))))))..(((((...((..(((((......)))))...))...)))))..................................................................................................................................... ........................................41...........................................................................118.................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................GCAGTGACGTCTCAGCAGTTACAGGAA........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTAGATGTCTGTCCCACAGG................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGACTGCTTTCCTGCGACGTTTCTTC.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |