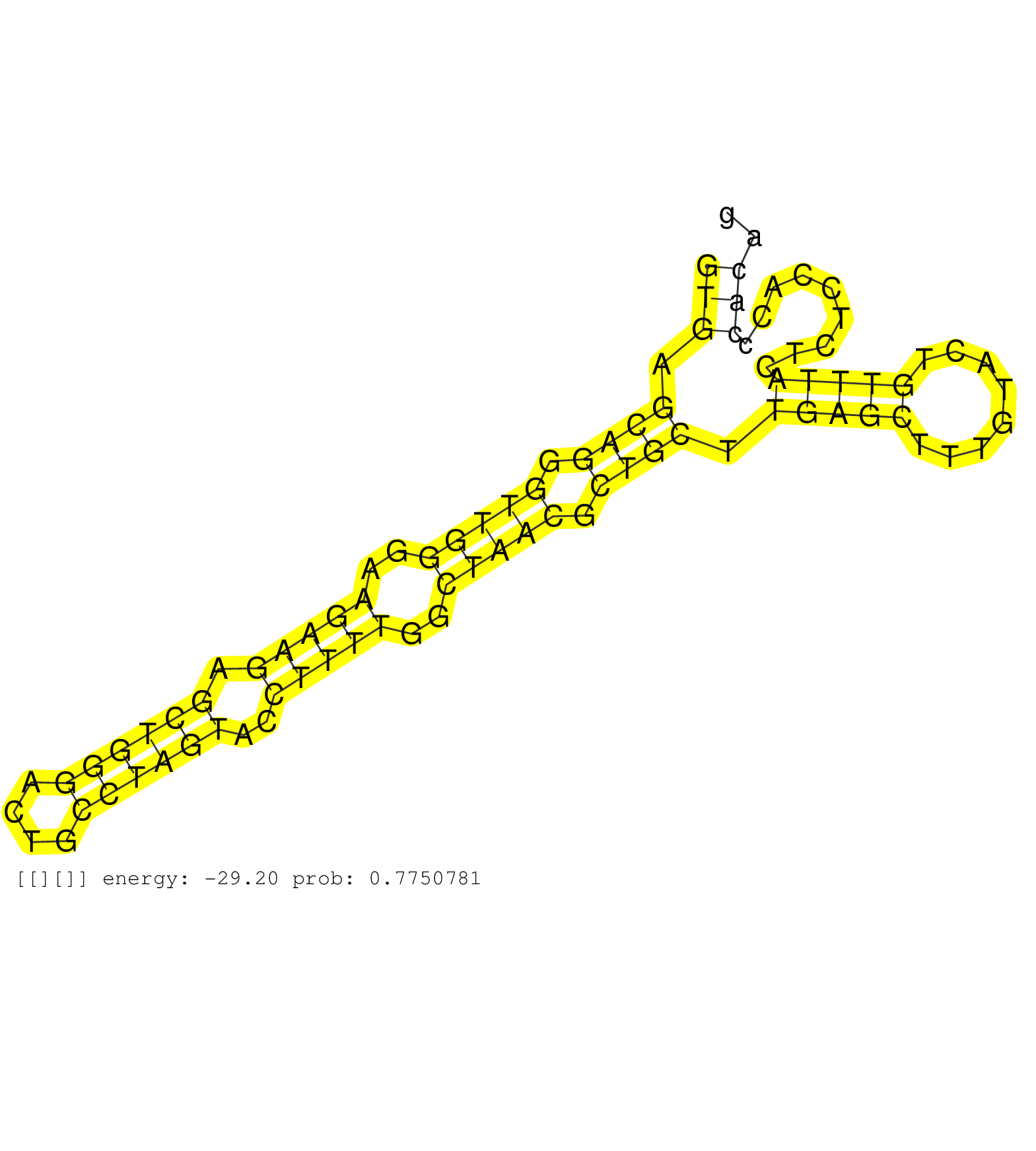

| Gene: Atxn2l | ID: uc009jrr.1_intron_5_0_chr7_133637844_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| TGTATCCAATTCCGTGCCTGGGCAGCAGGGCAAGTACCGGGGTGCAAAAGGTGAGCAGGGTTGGGAAGAAGAGCTGGGACTGCCTAGTACCTTTTGGCTAACGCTGCTTGAGCTTTGTACTGTTTACTCTCCACCCACAGGCTCACTTCCTCCTCAGCGCTCTGACCAACACCAACCAGCCTCAGCCCCT ..................................................(((.((((.(((((..(((((.((((((....))))))..)))))..))))).)))).(((((........))))).........))).................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGCAGGGTTGGGAAGAAGAGCTGGGACTGCCTAGTACCTTTTGGCTAAC........................................................................................ | 52 | 1 | 73.00 | 73.00 | 73.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGGGTTGGGAAGAAGAGC.................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................AGTACCGGGGTGCAAAAGGctca....................................................................................................................................... | 23 | ctca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................AGCGCTCTGACCAACACCAACCAGCCTC....... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............GTGCCTGGGCAGCAGGGCAAGT........................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CTCAGCGCTCTGACCAACACCA................ | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CAGGGCAAGTACCGGGGTGCAAAAG............................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................GCAGGGCAAGTACCGGGGTGCA................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TCAGCGCTCTGACCAACACCAACCAGC.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGACCAACACCAACCAGCCTCAG..... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................AAGTACCGGGGTGCAAAAGGctc........................................................................................................................................ | 23 | ctc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................GGAAGAAGAGCTGGcatc............................................................................................................. | 18 | catc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGGGTTGGGAAGAAGAGCTGGGAa.............................................................................................................. | 30 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TCAGCGCTCTGACCAACACCAAC.............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................CAGCGCTCTGACCAACACCAACC............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| TGTATCCAATTCCGTGCCTGGGCAGCAGGGCAAGTACCGGGGTGCAAAAGGTGAGCAGGGTTGGGAAGAAGAGCTGGGACTGCCTAGTACCTTTTGGCTAACGCTGCTTGAGCTTTGTACTGTTTACTCTCCACCCACAGGCTCACTTCCTCCTCAGCGCTCTGACCAACACCAACCAGCCTCAGCCCCT ..................................................(((.((((.(((((..(((((.((((((....))))))..)))))..))))).)))).(((((........))))).........))).................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................CGCTCTGACCAACtgg.................... | 16 | tgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCCTCCTCAGCGCTCccgc............................ | 19 | ccgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |