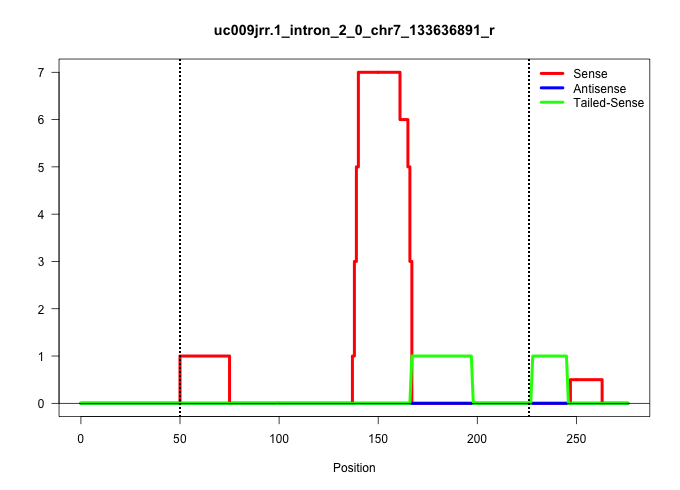
| Gene: Atxn2l | ID: uc009jrr.1_intron_2_0_chr7_133636891_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(10) TESTES |
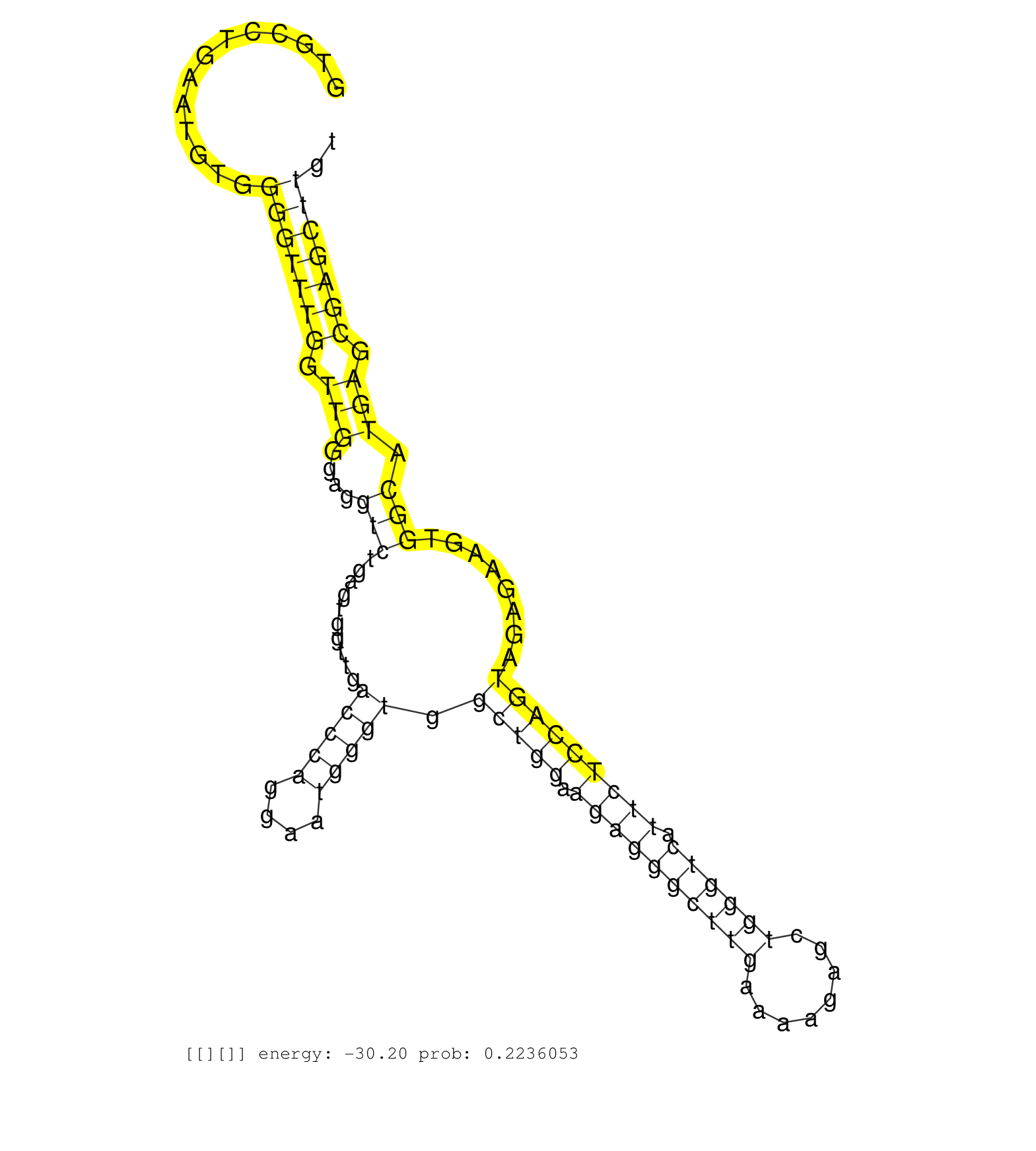
| CTACTGGGAGCCAGCCACAATCCCAGCATGCGGCCCCCAGTCCCGTTCAGGTGCCTGAATGTGGGGTTTGGTTGGGAGGTCTGAGTGGTTGACCCAGGAATGGGTGGCTGGAAGAGGGCTTGAAAAGAGCTGGGTCATTCTCCAGTAGAGAAGTGGCATGAGCGAGCTTGTGCTTTGTCTGCATAGCAGAATGGGTGGCACTCATACCTCACTTCTACTCTGGCAGCACCAGGCGGGGCAGGCCCCTCACCTGGGCAGTGGACAGCCACAGCAGAA ...............................................................(((((((.(((....(((..........(((((....))))).(((((.((((((((((........)))))).)))))))))........))).))).)))))))........................................................................................................... ..................................................51......................................................................................................................171....................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................TCCAGTAGAGAAGTGGCATGAGCGAGC............................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ....................................................................................................TGGGTGGCTGGAAGAGGGC............................................................................................................................................................. | 19 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CTCCAGTAGAGAAGTGGCATGA................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................TTCTCCAGTAGAGAAGTGGCATGAGCGA............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................TTCAGGTGCCTGAATGTGGGG.................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................CTCCAGTAGAGAAGTGGCATGAGCGAG.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGCCTGAATGTGGGGTTTGGTTGG......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................TTGTGCTTTGTCTGCATAGCAGAATGGGTGt.............................................................................. | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................TCTCCAGTAGAGAAGTGGCATGAGCGAGC............................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................TCTCCAGTAGAGAAGTGGCATGAGCGAG.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................CCAGGCGGGGCAGGgaac.............................. | 18 | gaac | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................................................CACCTGGGCAGTGGAC............. | 16 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................TGGACAGCCACAGCAGAA | 18 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - |
| CTACTGGGAGCCAGCCACAATCCCAGCATGCGGCCCCCAGTCCCGTTCAGGTGCCTGAATGTGGGGTTTGGTTGGGAGGTCTGAGTGGTTGACCCAGGAATGGGTGGCTGGAAGAGGGCTTGAAAAGAGCTGGGTCATTCTCCAGTAGAGAAGTGGCATGAGCGAGCTTGTGCTTTGTCTGCATAGCAGAATGGGTGGCACTCATACCTCACTTCTACTCTGGCAGCACCAGGCGGGGCAGGCCCCTCACCTGGGCAGTGGACAGCCACAGCAGAA ...............................................................(((((((.(((....(((..........(((((....))))).(((((.((((((((((........)))))).)))))))))........))).))).)))))))........................................................................................................... ..................................................51......................................................................................................................171....................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|