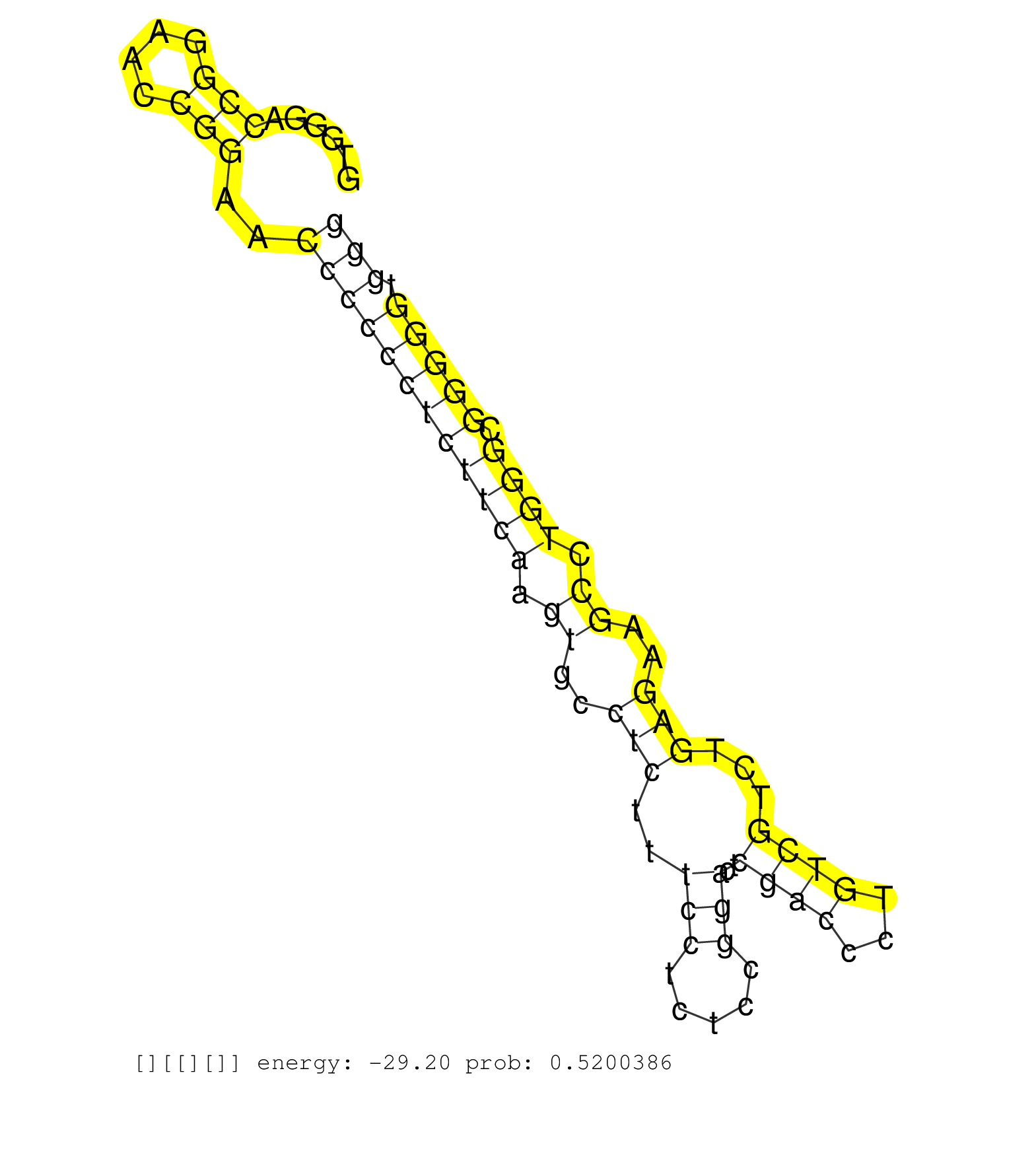
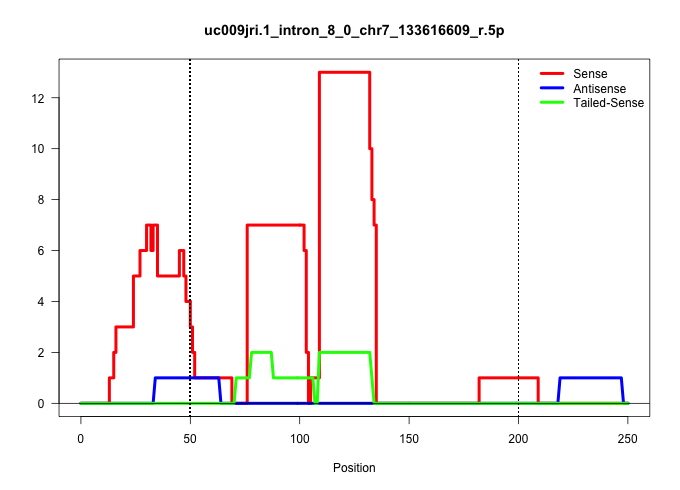
| Gene: Sh2b1 | ID: uc009jri.1_intron_8_0_chr7_133616609_r.5p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(5) PIWI.ip |
(4) PIWI.mut |
(27) TESTES |
| CGCCGCCGCCGCTGCCGCCGCCGGAGCTAAGCGCGGGGACCGAGATGCAGGTGGGACCGGAACCGGAACCCCCCTCTTCAAGTGCCTCTTTCCTCTCCGGACTCGACCCTGTCGTCTGAGAAGCCTGGGCGGGGGTGGGCGTGGGGGCTCGGGGCTGGAGGGGTGCTCGGCTCCCGAGAGGGCAGCAGATAGTTGGTCCCTCGGAGGCTCCGGCCCAGGGGCTTGTGGAGGGTGTCCAACGACTAGGATC ........................................................(((....)))..((((((((((((.((..(((..(((.....)))..((((...))))...)))..)).)))).))))).)))............................................................................................................... ..................................................51......................................................................................139............................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................TGTCGTCTGAGAAGCCTGGGCGGGGG................................................................................................................... | 26 | 1 | 8.00 | 8.00 | - | - | 4.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................TTCAAGTGCCTCTTTCCTCTCCGGACT................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CAGGGGCTTGTGGAGGGTGTa.............. | 21 | a | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTCGTCTGAGAAGCCTGGGCGG...................................................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................TTCAAGTGCCTCTTTCCTCTCCGGACTC.................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTCGTCTGAGAAGCCTGGGCGGG..................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GCCGCCGCCGGAGCTAAGC.......................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CGGGGACCGAGATGCAGG....................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCAGGTGGGACCGGAACCGGAAC..................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGGGCTTGTGGAGGGTGTCC............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CCCTGTCGTCTGAGAAGCCTGGGCGG...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............CGCCGCCGGAGCTAAGCGCG....................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CAAGTGCCTCTTTCCTCTCCGGACTCGct............................................................................................................................................... | 29 | ct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGCTAAGCGCGGGGACCGAGATGC.......................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................CCCTCTTCAAGTGagag.................................................................................................................................................................. | 17 | agag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................ACCCTGTCGTCTGAGAAGCCTGGGCGG...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTCGTCTGAGAAGCCTGGGCGGGt.................................................................................................................... | 25 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTCGTCTGAGAAGCCTGGGCGGGG.................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CAGCAGATAGTTGGTCCCTCGGAGGCT......................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TAAGCGCGGGGACCGAGATGCAGGT...................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CAGATAGTTGGTCCCTCGGAG............................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TTCAAGTGCCTCTTTCCTCTCCGGAC.................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTCGTCTGAGAAGCCTGGGCGGa..................................................................................................................... | 24 | a | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................CTCCGGACTCGACCCTGTC......................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................GCGCGGGGACCGAGATGCAG........................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................AGCTAAGCGCGGGGACCGAGATG........................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GCCGCCGGAGCTAAGCGCG....................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................GGGCTCGGGGCTGGAG.......................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| CGCCGCCGCCGCTGCCGCCGCCGGAGCTAAGCGCGGGGACCGAGATGCAGGTGGGACCGGAACCGGAACCCCCCTCTTCAAGTGCCTCTTTCCTCTCCGGACTCGACCCTGTCGTCTGAGAAGCCTGGGCGGGGGTGGGCGTGGGGGCTCGGGGCTGGAGGGGTGCTCGGCTCCCGAGAGGGCAGCAGATAGTTGGTCCCTCGGAGGCTCCGGCCCAGGGGCTTGTGGAGGGTGTCCAACGACTAGGATC ........................................................(((....)))..((((((((((((.((..(((..(((.....)))..((((...))))...)))..)).)))).))))).)))............................................................................................................... ..................................................51......................................................................................139............................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........CCGCCGCCGGAGCgcgc............................................................................................................................................................................................................................... | 17 | gcgc | 16.00 | 0.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTCCTCTCCGGACTCagc.................................................................................................................................................. | 19 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................GGGGACCGAGATGCAGGTGGGACCGGAACC.......................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......GCTGCCGCCGCCGGcaac.................................................................................................................................................................................................................................. | 18 | caac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................GGCTTGTGGAGGGTGTCCAACGACTAGGA.. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |