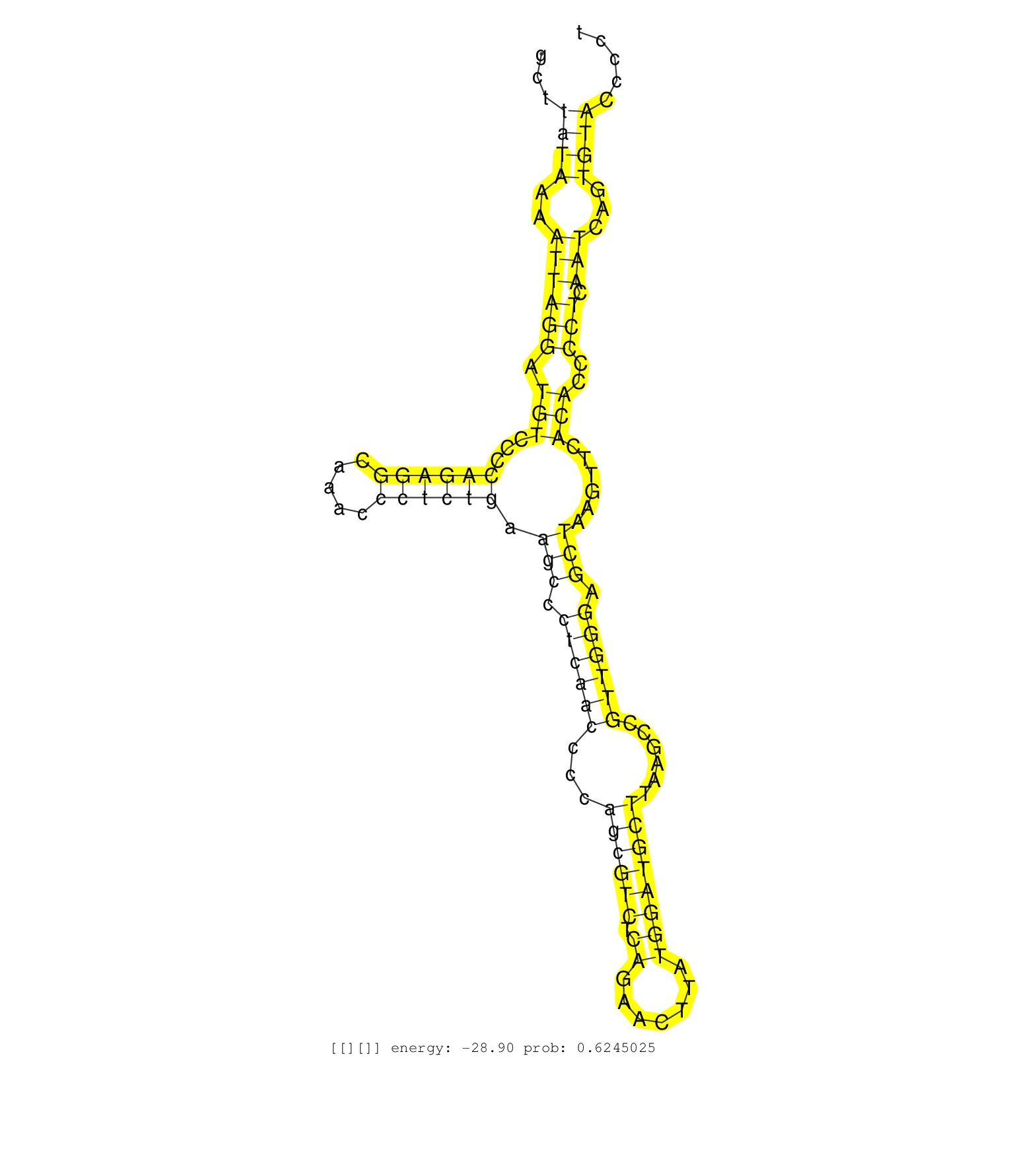

| Gene: Spns1 | ID: uc009jqy.1_intron_6_0_chr7_133516405_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| AGAACAATCAGGGTCCTGTCACCCACCAAAGAAGCAGCAATGTCAGGCTTATAAAATTAGGATGTCCCCAGAGGCAAACCCTCTGAAGCCCTCAACCCCAGCGTCTCAGAACTTATGGATGCTTAAGCCGTTGGGAGCTAAGTTCACACCCCTCAATCAGTGTACCCCTGCCCGTGTCCCAAAGCTTGCACTTTTCCCAGGTGACACCAGGTCTAGGAGTGCTGGCTGTCCTGCTGCTGTTCCTGGTGGT .................................................((((..((((((.(((...((((((.....)))))).(((.((((((...((((((.((.......))))))))......)))))).)))......)))..))).)))...))))...................................................................................... ..............................................47........................................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGTTGGGAGCTAAGTTCACACCCCTC................................................................................................ | 52 | 1 | 89.00 | 89.00 | 89.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGT....................................................................................................................... | 29 | 1 | 28.00 | 28.00 | - | 23.00 | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGTT...................................................................................................................... | 30 | 1 | 17.00 | 17.00 | - | 13.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CGTCTCAGAACTTATGGATGCTTAAGCCGT....................................................................................................................... | 30 | 1 | 6.00 | 6.00 | - | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................TCTCAGAACTTATGGATGCTTAAGCCGTT...................................................................................................................... | 29 | 1 | 6.00 | 6.00 | - | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCTCAGAACTTATGGATGCTTAAGCCG........................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................TCAGAACTTATGGATGCTTAAGCCGTT...................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCTCAGAACTTATGGATGCTTAAGCCGT....................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCTAGGAGTGCTGGCTGTC.................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AGAACTTATGGATGCTTAAGCCGTTG..................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGacgt....................................................................................................................... | 29 | acgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGa....................................................................................................................... | 29 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCTCAGAACTTATGGATGCTTAAac.......................................................................................................................... | 25 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................TCAGAACTTATGGATGCTTAAGCCGTTG..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCcttg..................................................................................................................... | 31 | cttg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TGGGAGCTAAGTTCACACCCCTCAATCAGT......................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................TGGATGCTTAAGCCGTTGGGAGCTAAGTTC......................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATG................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGgcgt....................................................................................................................... | 29 | gcgt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGTg...................................................................................................................... | 30 | g | 1.00 | 28.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TCAGAACTTATGGATagt............................................................................................................................... | 18 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................GAACTTATGGATGCTTAAGCCGTTG..................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGg....................................................................................................................... | 29 | g | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTATGGATGCTTAAGCCGTTGGGAGCT............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCTCAGAACTTATGGATGCTTAAGCCGat...................................................................................................................... | 29 | at | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAAATTAGGATGTCCCCAGAGGC............................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................GTCTCAGAACTTATGGATGCTTAAGCCGgt...................................................................................................................... | 30 | gt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CGTCTCAGAACTTATGGATGCTTAAGacgt....................................................................................................................... | 30 | acgt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAACAATCAGGGTCCTGTCACCCACCAAAGAAGCAGCAATGTCAGGCTTATAAAATTAGGATGTCCCCAGAGGCAAACCCTCTGAAGCCCTCAACCCCAGCGTCTCAGAACTTATGGATGCTTAAGCCGTTGGGAGCTAAGTTCACACCCCTCAATCAGTGTACCCCTGCCCGTGTCCCAAAGCTTGCACTTTTCCCAGGTGACACCAGGTCTAGGAGTGCTGGCTGTCCTGCTGCTGTTCCTGGTGGT .................................................((((..((((((.(((...((((((.....)))))).(((.((((((...((((((.((.......))))))))......)))))).)))......)))..))).)))...))))...................................................................................... ..............................................47........................................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................CCTGCCCGTGTCCCAAAGCTTGCACT.......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GCCCTCAACCCCAGCGTCTCAGAACTTA....................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |