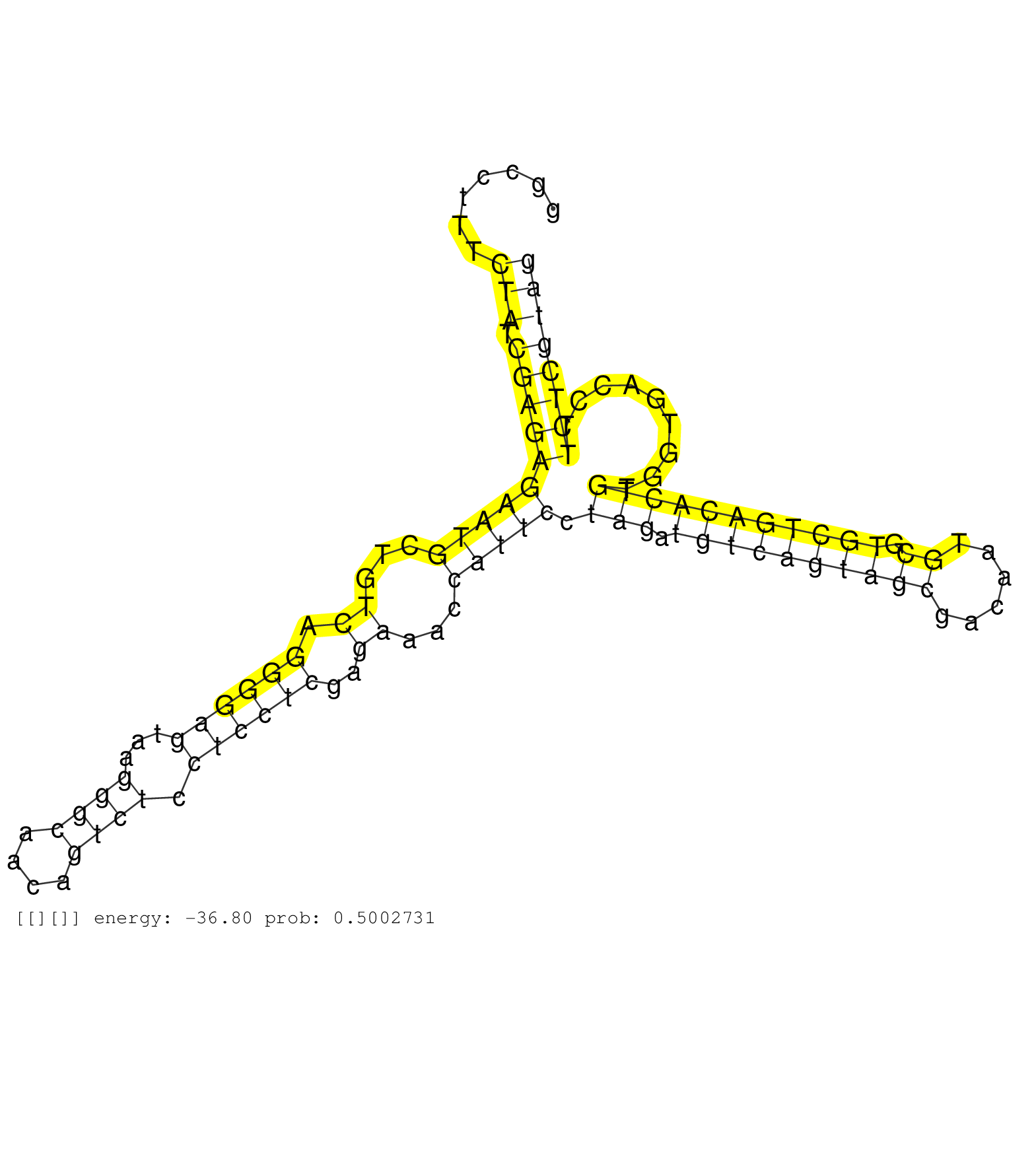

| Gene: Jmjd5 | ID: uc009jpz.1_intron_2_0_chr7_132598772_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OVARY |
(6) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| CTAAGGGTTGGGCAGGCGAGGGCGGCCTTCCAGAGCCATATGCAAAGCCACCTCAGCTGCATGGTCTCAGGCACACAGCCTGGCCTTTCTATCGAGAGAATGCTGTCAGGGGAGTAAGGGCAACAGTCTCCTCCTCGAGAAACCATTCCTAGATGTCAGTAGCGACAATGCGTGCTGACACTGTGGTGACCTTCTCGTAGTCTGCAGTACATCCAGGAGATTGCTGGCTGCCGCACCGTCCCCGTGGAAG ........................................................................................(((.((((((((((...((.((((((...((((....)))).))))))..))...))))).(((.((((((((((......)).))))))))))).........)))))))).................................................. .................................................................................82....................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TTCTATCGAGAGAATGCTGTCAGGGG.......................................................................................................................................... | 26 | 1 | 9.00 | 9.00 | 4.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................TTTCTATCGAGAGAATGCTGTCAGGGG.......................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | 3.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTCTATCGAGAGAATGCTGTCAGGG........................................................................................................................................... | 26 | 1 | 6.00 | 6.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCGTAGTCTGCAGTACATCCAGGAGA.............................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TCTCGTAGTCTGCAGTACATCCAGGAG............................... | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTCTATCGAGAGAATGCTGTCAGGG........................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGCGTGCTGACACTGTGGTGACCTTCTC...................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAAACCATTCCTAGATGTCAGTAGCGACA................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CGTGCTGACACTGTGGTGACCTTCTCGTAGaa................................................ | 32 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................TTCTATCGAGAGAATGCTGTCAGGGGAGT....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CGAGAAACCATTCCTAGATGTCAGTAG........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCGTAGTCTGCAGTACATCCAGGAGAT............................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GGAGATTGCTGGCTGCCGCACCGTCCCt....... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCGTAGTCTGCAGTACATCCAGGAG............................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTCTATCGAGAGAATGCTGTCAGG............................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTCCTAGATGTCAGTAGCGACAATGCGTG............................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TATCGAGAGAATGCTGTCAGGGGAGT....................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TACATCCAGGAGATTGCTGGCTGCC.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................TCGTAGTCTGCAGTACATCCAGGAGATT............................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................TGCGTGCTGACACTGTGGTGACCTTCT....................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TCTATCGAGAGAATGCTGTCAGGGGA......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTCCTAGATGTCAGTAGCGACAATGCGTGC........................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CTAAGGGTTGGGCAGGCGAGGGCGGCCTTCCAGAGCCATATGCAAAGCCACCTCAGCTGCATGGTCTCAGGCACACAGCCTGGCCTTTCTATCGAGAGAATGCTGTCAGGGGAGTAAGGGCAACAGTCTCCTCCTCGAGAAACCATTCCTAGATGTCAGTAGCGACAATGCGTGCTGACACTGTGGTGACCTTCTCGTAGTCTGCAGTACATCCAGGAGATTGCTGGCTGCCGCACCGTCCCCGTGGAAG ........................................................................................(((.((((((((((...((.((((((...((((....)))).))))))..))...))))).(((.((((((((((......)).))))))))))).........)))))))).................................................. .................................................................................82....................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................TTCTCGTAGTCTGCAGTACATCCAGGA................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................TGCAAAGCCACCTCAGCTGCATGGTC........................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................GGTGACCTTCTCGTAGTCTGCAGTACA....................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................TCTCGTAGTCTGCAGTACATCCAGGA................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................CTTCTCGTAGTCTGCAGTACATCCA................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GACCTTCTCGTAGTCTGCAGTACATCCA................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |