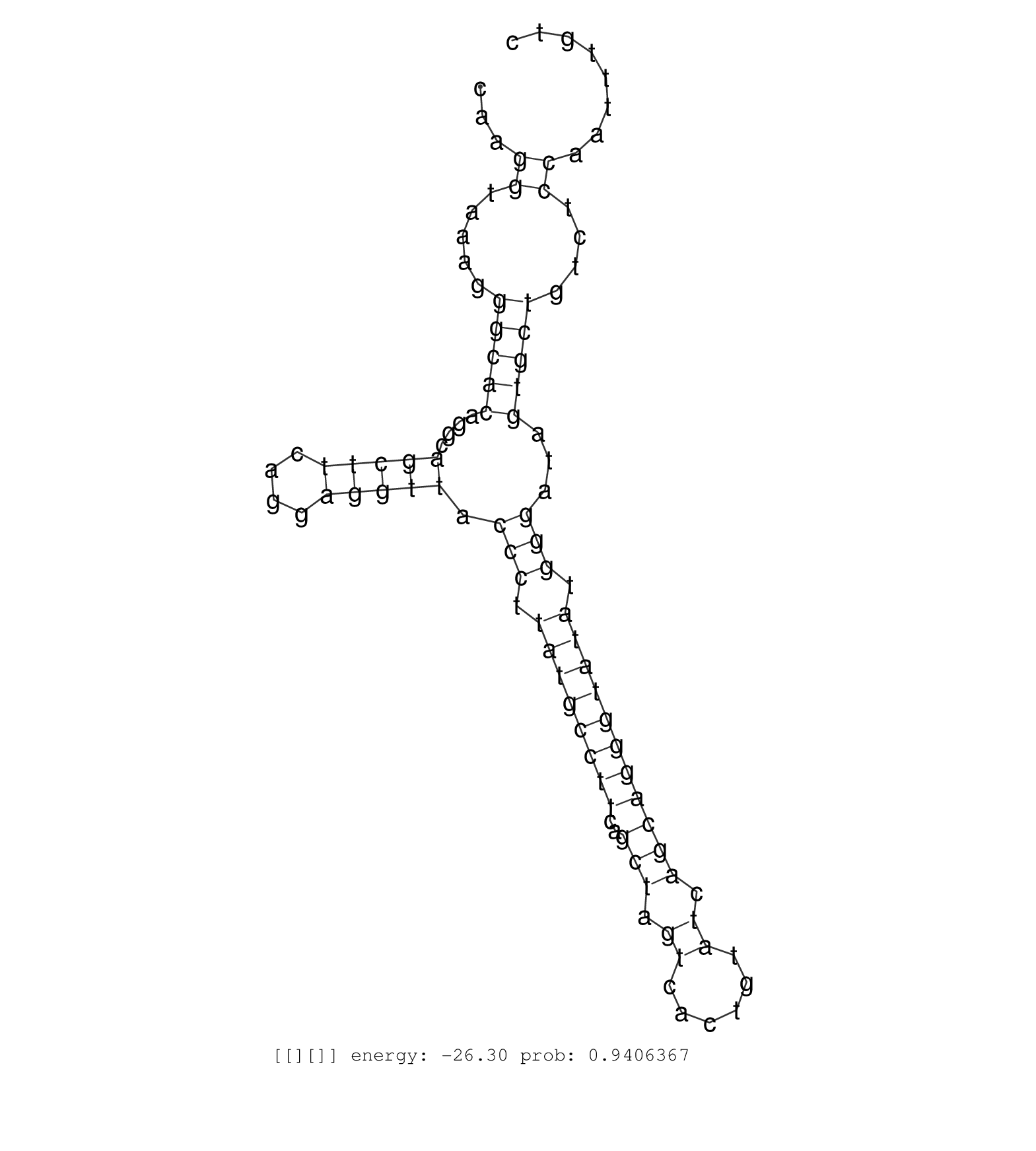

| Gene: Plk1 | ID: uc009joo.1_intron_1_0_chr7_129277698_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(4) PIWI.mut |
(21) TESTES |
| CTGTTTTTTCACTCCAGATTTCTGTTTGCTTTTAGGCATCTGGGAACAAAGTTAGTCGCCAGTCTGTTTTGTGTGGAAGCCAGAACATCGTCCTCAATGGCAAGGTAAAGGGCACAGGCAGCTTCAGGAGGTTACCCTTATGCCTTCAGCTAGTCACTGTATCAGCAGGGTATATGGGATAGTGCTGTCTCCAATTTGTCATTTCCAGTTCTCTCTCTTTGTTTTTGTTTTTTTAAACATTTATTTATTT .......................................................................................................((.....(((((....(((((....))))).(((.((((((((..(((.((......)).))))))))))).)))...)))))....)).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................TTTGTGTGGAAGCCAGAACATCGTCCTC........................................................................................................................................................... | 28 | 1 | 13.00 | 13.00 | 4.00 | 2.00 | - | 2.00 | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................CCAGAACATCGTCCTCAATGGCAAGac................................................................................................................................................ | 27 | ac | 5.00 | 0.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTTGTGTGGAAGCCAGAACATCGTCCT............................................................................................................................................................ | 27 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGTGGAAGCCAGAACATCGTCCTC........................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTTGTGTGGAAGCCAGAACATCGTCC............................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TAGTCGCCAGTCTGTTTTGTGTGGA............................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGTGGAAGCCAGAACATCGTCCTCA.......................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................AGAACATCGTCCTCAATGGCAtc.................................................................................................................................................. | 23 | tc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGTGGAAGCCAGAACATCGTCC............................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................TGGAAGCCAGAACATCGTCCTCAATGGC..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................AGCCAGAACATCGTCCTC........................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................ATCGTCCTCAATGGCAAGacca.............................................................................................................................................. | 22 | acca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................CAGAACATCGTCCTCAATGGCAAGacc............................................................................................................................................... | 27 | acc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................TGGAAGCCAGAACATCGTCCT............................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................AGTTAGTCGCCAGTCTGTTTTGTG................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................AAGTTAGTCGCCAGTCTGTTTTGTG................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................TGGAAGCCAGAACATCGTCCTCAATGGCAA................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TTTTGTGTGGAAGCCAGAACATCGTCCT............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGTGGAAGCCAGAACATCGTCCTCAAT........................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGAAGCCAGAACATCGTCCTCAATGt...................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................AACAAAGTTAGTCGCCAGTCTGTTTT.................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................TTGTGTGGAAGCCAGAACATCGTCCT............................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGTGGAAGCCAGAACATCGTCCT............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................AGCCAGAACATCGTCCT............................................................................................................................................................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGTTTTTTCACTCCAGATTTCTGTTTGCTTTTAGGCATCTGGGAACAAAGTTAGTCGCCAGTCTGTTTTGTGTGGAAGCCAGAACATCGTCCTCAATGGCAAGGTAAAGGGCACAGGCAGCTTCAGGAGGTTACCCTTATGCCTTCAGCTAGTCACTGTATCAGCAGGGTATATGGGATAGTGCTGTCTCCAATTTGTCATTTCCAGTTCTCTCTCTTTGTTTTTGTTTTTTTAAACATTTATTTATTT .......................................................................................................((.....(((((....(((((....))))).(((.((((((((..(((.((......)).))))))))))).)))...)))))....)).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................CACAGGCAGCTTCAGtctg........................................................................................................................... | 19 | tctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |