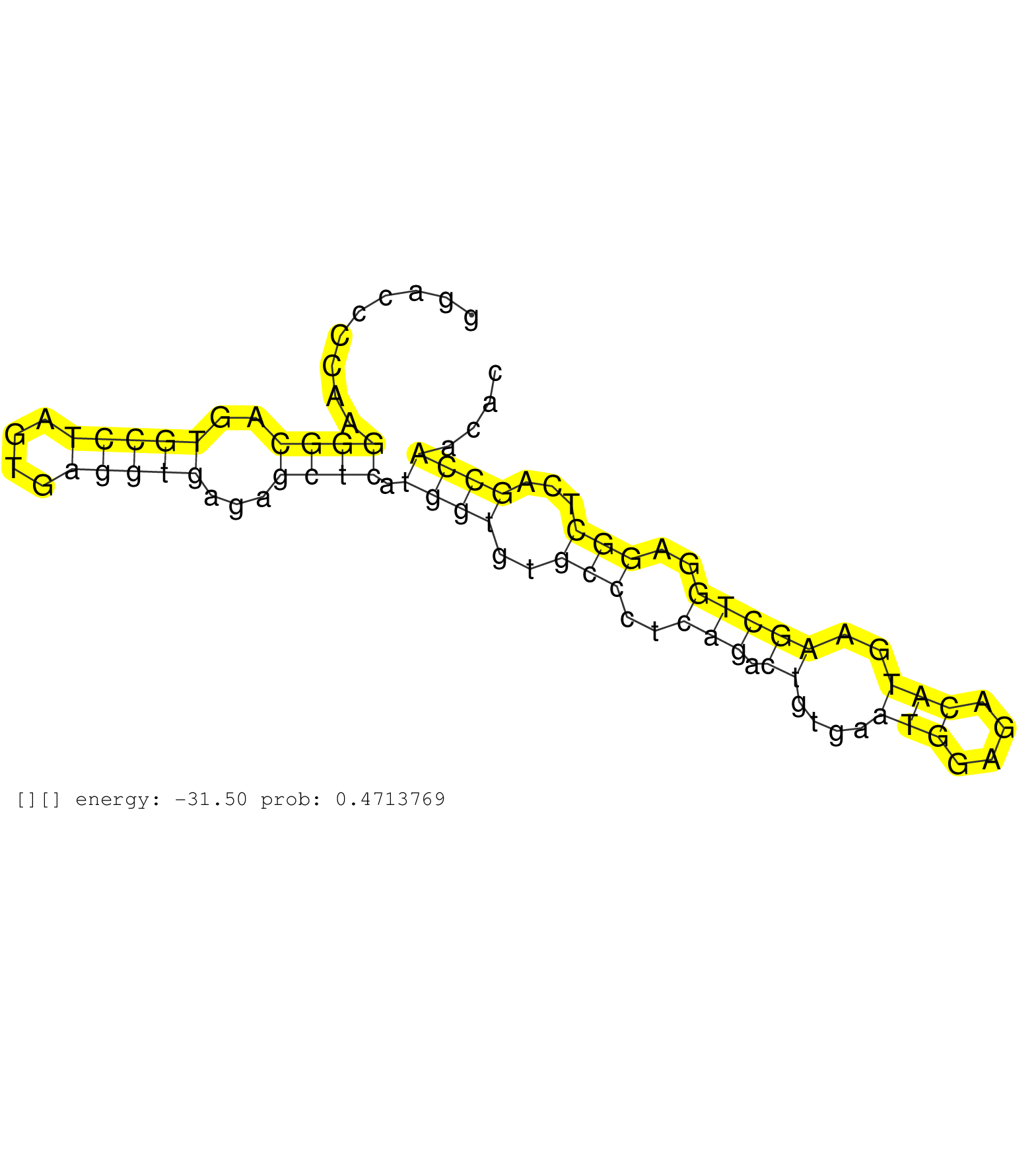

| Gene: AK016693 | ID: uc009jia.1_intron_2_0_chr7_121464766_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| GAGGGACTGATCACAGTCCTGAGACGCCCCGAAGCACAGTCCCCTTTGAGGTAAGTCACATACAAGACCCACTCAGGTCACTGCGAGGTCTTGGGGAGACATGGACCCCAAGGGCAGTGCCTAGTGAGGTGAGAGCTCATGGTGTGCCCTCAGACTGTGAATGGAGACATGAAGCTGGAGGCTCAGCCAACACTGAGGCCAGACCAGAATGAAGAAACTCAACACTGTCAGTCACATCTGGCTTCCAGGG ...............................................................................................................((((..(((((....)))))...)))).((((..(((..(((.((....(((....)))..)))))..)))...))))............................................................. ......................................................................................................103.......................................................................................193....................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................TGGAGACATGAAGCTGGAGGCTCAGCCA............................................................. | 28 | 1 | 9.00 | 9.00 | 1.00 | 2.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGGAGACATGAAGCTGGAGGCTCAGCCAA............................................................ | 29 | 1 | 6.00 | 6.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGTGAATGGAGACATGAAGCTGGAGGCTC.................................................................. | 29 | 1 | 4.00 | 4.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAAGAAACTCAACACTGTCAGTCACATC............ | 29 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CTCAGACTGTGAATGGAGACATGAAG............................................................................ | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGTGAATGGAGACATGAAGCTGGAGGCT................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGAATGGAGACATGAAGCTGGAGGCTC.................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCCCTCAGACTGTGAATGGAGACATGA.............................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGTGAATGGAGACATGAAGCTGGAGG..................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................TCAGACTGTGAATGGAGACATGAAGCTGt........................................................................ | 29 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGAATGGAGACATGAAGCTGGAGGCTCA................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TCAGACTGTGAATGGAGACATGAAGCTG......................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TCAGACTGTGAATGGAGACATGAAGCTGGA....................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................TCAGACTGTGAATGGAGACATGAAGC........................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......TGATCACAGTCCTGAGACGCCCCGAAG........................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGATCACAGTCCTGAGACGCCCCGAAGC....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................TGGAGACATGAAGCTGGAGGCTCAGCC.............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................CCAAGGGCAGTGCCTgtga............................................................................................................................ | 19 | gtga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................................TGAGGCCAGACCAGAATGAAGAAACTCAc............................ | 29 | c | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGGAGACATGAAGCTGGAGGCTCAGt............................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................TGAGGTAAGTCACATACAAGACCCACT................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................CTGTGAATGGAGACATGAAGCTGGA....................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GAATGGAGACATGAAGCTGGAGGCTCA................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAAGAAACTCAACACTGTCAGTCACATCT........... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GAGGGACTGATCACAGTCCTGAGACGCCCCGAAGCACAGTCCCCTTTGAGGTAAGTCACATACAAGACCCACTCAGGTCACTGCGAGGTCTTGGGGAGACATGGACCCCAAGGGCAGTGCCTAGTGAGGTGAGAGCTCATGGTGTGCCCTCAGACTGTGAATGGAGACATGAAGCTGGAGGCTCAGCCAACACTGAGGCCAGACCAGAATGAAGAAACTCAACACTGTCAGTCACATCTGGCTTCCAGGG ...............................................................................................................((((..(((((....)))))...)))).((((..(((..(((.((....(((....)))..)))))..)))...))))............................................................. ......................................................................................................103.......................................................................................193....................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|