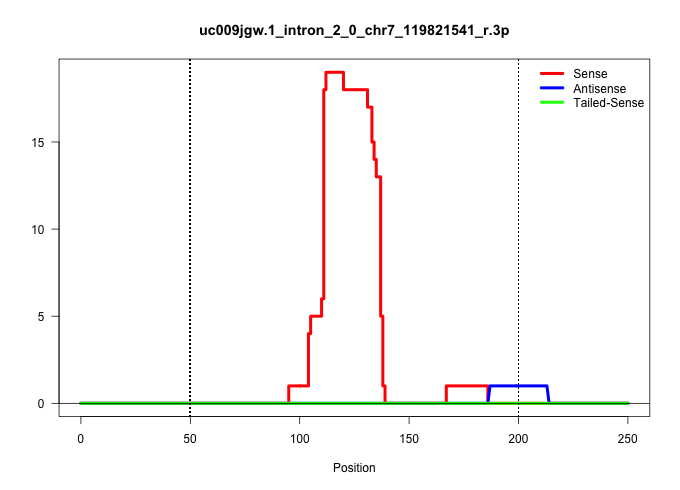
| Gene: AK009335 | ID: uc009jgw.1_intron_2_0_chr7_119821541_r.3p | SPECIES: mm9 |
 |
|
(5) OTHER.mut |
(2) PIWI.mut |
(14) TESTES |
| TTTTTTAATTCTATACATTCTATAATTTCTCACCTGTGTAGATGTCCACCGCAATCAGGTACAAATGTGTATTATTTCCAGTTACTATGCAGTAATGTGACAGGTTGACAGTAAGAACTTCACACACCCGAGCAGAGTATTAGAAAATACACGACGATTTTTCCCTTTAAAACTCTGGCAGGTGTATGTCTCTTTTCCAGATAGCCTTCCATTAACACCAGCGAATAAGACAGAGGGCAGAAAACGCCAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TAAGAACTTCACACACCCGAGCAGAG................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................TAAGAACTTCACACACCCGAGCAGAGT................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................TTGACAGTAAGAACTTCACACACCCGAGC..................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGTGACAGGTTGACAGTAAGAACTT.................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................TGACAGTAAGAACTTCACACACCCGAGCA.................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTAAGAACTTCACACACCCGAGCAG................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................AAGAACTTCACACACCCGAGCAGAGT................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................TTGACAGTAAGAACTTCACACACCCGA....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................TAAAACTCTGGCAGGTGTA................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................TAAGAACTTCACACACCCGAGCAGAGTA............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| TTTTTTAATTCTATACATTCTATAATTTCTCACCTGTGTAGATGTCCACCGCAATCAGGTACAAATGTGTATTATTTCCAGTTACTATGCAGTAATGTGACAGGTTGACAGTAAGAACTTCACACACCCGAGCAGAGTATTAGAAAATACACGACGATTTTTCCCTTTAAAACTCTGGCAGGTGTATGTCTCTTTTCCAGATAGCCTTCCATTAACACCAGCGAATAAGACAGAGGGCAGAAAACGCCAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................GTCTCTTTTCCAGATAGCCTTCCATTA.................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................CGCAATCAGGTACAaacc........................................................................................................................................................................................... | 18 | aacc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |