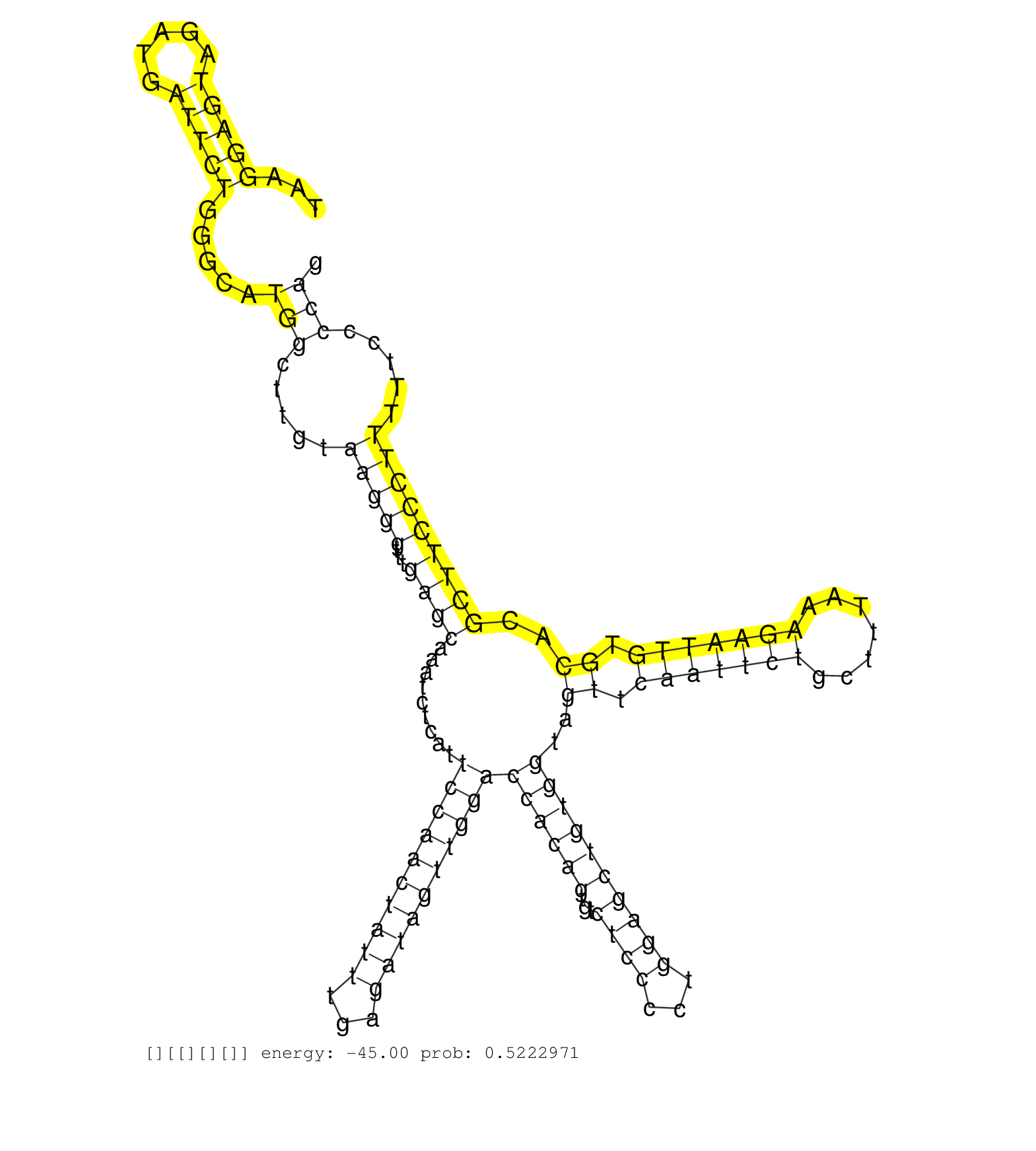

| Gene: Trpc2 | ID: uc009iql.1_intron_9_0_chr7_109230661_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| ACCCTCTTCCTACTCACATGGTTCTTAAGGGAGCCAGAGAGATTCCTTTCTAAGGAGTAGATGATTCTGGGCATGGCTTGTAAGGGTTTGAGCAAATCTCATTCCAACTATTTGAGATAGTTGGACCACAGTGTCTCCCCTGGAGCTGTGGTAGTTCAATTCTGCTTTAAAGAATTGTGCACGCTTCCCTTTTTCCCCAGGAGACGTCATCATGTGGCACTAACACCCGTTCCCCTTGTCCCCAAGGTAC .....................................................(((((.....))))).....(((.....(((((...((((.........((((((((((...))))))))))((((((...((((...))))))))))..((.(((((((.......))))))).))..))))))))).....)))................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TAAGGAGTAGATGATTCTGGGCATG............................................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................TAAAGAATTGTGCACGCTTCCCTTTT......................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................AGATGATTCTGGGCATGGCTTGTAAGG..................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................CTAAGGAGTAGATGATTCTGGGCATG............................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................................CCCAGGAGACGTCATCATGTGGCACTAt........................... | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GATAGTTGGACCACAGTGTCTCC................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTTGAGATAGTTGGACCACAGTGTCTCC................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................CTAAGGAGTAGATGATTCTGGGCATGGCT............................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................AAGGAGTAGATGATTCTGGGCATGGCT............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTTGAGATAGTTGGACCACAGTGTCT.................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GGAGTAGATGATTCTGGGCATGG.............................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TAAGGAGTAGATGATTCTGGGCATGGCT............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................CTAAGGAGTAGATGATTCTGGGCATGG.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................TCTAAGGAGTAGATGATTCTGGGCAT................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGAGACGTCATCATGTGGCACTAACACC....................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAGACGTCATCATGTGGCACTAACACC....................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TCTGGGCATGGCTTGTAAGG..................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TGAGATAGTTGGACCACAGTGTCTCCCC.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TAAAGAATTGTGCACGCTTCCCTct.......................................................... | 25 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................GATGATTCTGGGCATGGCTTGTAAGGG.................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................TAAGGAGTAGATGATTCTGGGCATGGCTa........................................................................................................................................................................... | 29 | a | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGAGACGTCATCATGTGGCACTAACAC........................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................TTCTAAGGAGTAGATGATTCTGGGC.................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................CCTTTCTAAGGAGTAGATGATTCTGGGC.................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GTAGATGATTCTGGccgc................................................................................................................................................................................ | 18 | ccgc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACCCTCTTCCTACTCACATGGTTCTTAAGGGAGCCAGAGAGATTCCTTTCTAAGGAGTAGATGATTCTGGGCATGGCTTGTAAGGGTTTGAGCAAATCTCATTCCAACTATTTGAGATAGTTGGACCACAGTGTCTCCCCTGGAGCTGTGGTAGTTCAATTCTGCTTTAAAGAATTGTGCACGCTTCCCTTTTTCCCCAGGAGACGTCATCATGTGGCACTAACACCCGTTCCCCTTGTCCCCAAGGTAC .....................................................(((((.....))))).....(((.....(((((...((((.........((((((((((...))))))))))((((((...((((...))))))))))..((.(((((((.......))))))).))..))))))))).....)))................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................TTTTCCCCAGGAGACggta............................................. | 19 | ggta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |