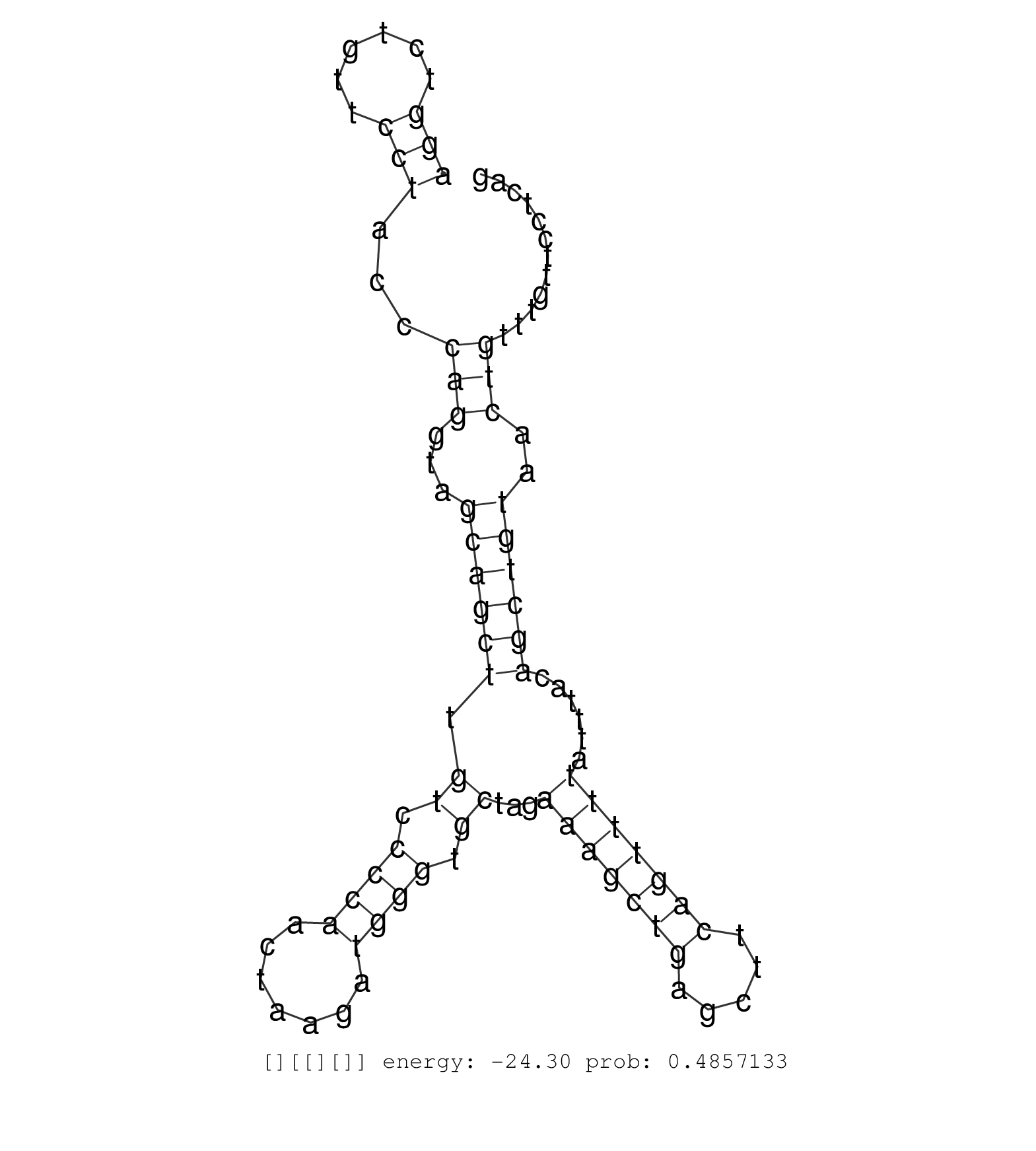

| Gene: Il18bp | ID: uc009iqd.1_intron_3_0_chr7_109165456_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(25) TESTES |
| CCAGACTTGCTTGCAAGTCATCATGACCATGAGACACTGCTGGACAGCAGGTGGGACATTAGGCTTTGTGGCTCTTAAAGGAATGGAGCTATGTGGCTAACTCTGCTACCCAACACCCCCCACCCCAAGGAAGTTCCATCCAATTCTTCCATTGCTGTAAGCGCCTATGTAGCTACTTTGGTTCTGGCTCCATTTCAAGGCTCCTTTGCACTACATTCAGCAGAGCCGAGGGGTGGGGGCTAAACGGTCTCCACCCACTCAGGTTCCTGGTGAGAACCTAGGAGTATCTGGGTCGAGGTCTGTTCCTACCCAGGTAGCAGCTTGTCCCCAACTAAGATGGGTGCTAGAAAGCTGAGCTTCAGTTTTATTTACAGCTGTAACTGTTTGTTCCTCAGGCCCCAGTTCTTGGTGGGTCCTGCTTTTGTATGTCCATGTCATTTTGGCC .......................................................................................................................................................................................................................................................................................................(((......)))...(((...((((((.((.((((.......)))).))...(((((((.....)))))))......))))))..))).............................................................. .......................................................................................................................................................................................................................................................................................................296................................................................................................395................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................TGACCATGAGACACTGCTGGACAGCAGGc......................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | c | 30.00 | 11.00 | 11.00 | - | 5.00 | 6.00 | 1.00 | - | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ..................................................GTGGGACATTAGGCTTTGTGGCTCTTAAAGGAATGGAGCTATGTGGCTAACT....................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 23.00 | 23.00 | - | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGcc........................................................................................................................................................................................................................................................................................................................................................................................................ | 30 | cc | 12.00 | 11.00 | 4.00 | - | 3.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGG.......................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 11.00 | 11.00 | 6.00 | - | 1.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGccc....................................................................................................................................................................................................................................................................................................................................................................................................... | 31 | ccc | 3.00 | 11.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................TCATCATGACCATGAGACACTGCTGGA................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGT......................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGcccc...................................................................................................................................................................................................................................................................................................................................................................................................... | 32 | cccc | 2.00 | 11.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGccct...................................................................................................................................................................................................................................................................................................................................................................................................... | 32 | ccct | 2.00 | 11.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GACCATGAGACACTGCTGGACAGCAGGcc........................................................................................................................................................................................................................................................................................................................................................................................................ | 29 | cc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................ATCATGACCATGAGACACTGCTGGAC................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................TTCTTGGTGGGTCCTGCTTTTGTATG................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........TGCAAGTCATCATGACCATGAGACACT....................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................ACCATGAGACACTGCTGGACAGCAGGcc........................................................................................................................................................................................................................................................................................................................................................................................................ | 28 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GACCATGAGACACTGCTGGACAGCAGGc......................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGcaa....................................................................................................................................................................................................................................................................................................................................................................................................... | 31 | caa | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGg......................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | g | 1.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ATGACCATGAGACACTGCTGGACAGgagg.......................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | gagg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TCATGACCATGAGACACTGCTGGACAGC............................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TCATGACCATGAGACACTGCTGGACAG.............................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACCATGAGACACTGCTGGACAGCAGGgc........................................................................................................................................................................................................................................................................................................................................................................................................ | 30 | gc | 1.00 | 11.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TCATCATGACCATGAGACACTGCTGGAC................................................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ACCATGAGACACTGCTGGACAGCAGGcccc...................................................................................................................................................................................................................................................................................................................................................................................................... | 30 | cccc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................ATCATGACCATGAGACACTGCTGGACAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................ATGAGACACTGCTGGACAGCAGGcc........................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | cc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TAAAGGAATGGAGCTATGTGGCTAACT....................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GAGGGGTGGGGGCTctcc........................................................................................................................................................................................................ | 18 | ctcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| CCAGACTTGCTTGCAAGTCATCATGACCATGAGACACTGCTGGACAGCAGGTGGGACATTAGGCTTTGTGGCTCTTAAAGGAATGGAGCTATGTGGCTAACTCTGCTACCCAACACCCCCCACCCCAAGGAAGTTCCATCCAATTCTTCCATTGCTGTAAGCGCCTATGTAGCTACTTTGGTTCTGGCTCCATTTCAAGGCTCCTTTGCACTACATTCAGCAGAGCCGAGGGGTGGGGGCTAAACGGTCTCCACCCACTCAGGTTCCTGGTGAGAACCTAGGAGTATCTGGGTCGAGGTCTGTTCCTACCCAGGTAGCAGCTTGTCCCCAACTAAGATGGGTGCTAGAAAGCTGAGCTTCAGTTTTATTTACAGCTGTAACTGTTTGTTCCTCAGGCCCCAGTTCTTGGTGGGTCCTGCTTTTGTATGTCCATGTCATTTTGGCC .......................................................................................................................................................................................................................................................................................................(((......)))...(((...((((((.((.((((.......)))).))...(((((((.....)))))))......))))))..))).............................................................. .......................................................................................................................................................................................................................................................................................................296................................................................................................395................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................CCAATTCTTCCATtaga..................................................................................................................................................................................................................................................................................................... | 17 | taga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....GCTTGCAAGTCATCAgag...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | gag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................AGGCTCCTTTGCACTACATTCAGCA............................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CTTGCAAGTCATCATGagg.................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................GTGGGTCCTGCTTTTGTATGTCCATGTCA........ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................................AACTGTTTGTTCCTCAGGCCCCAGTTCTT...................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |