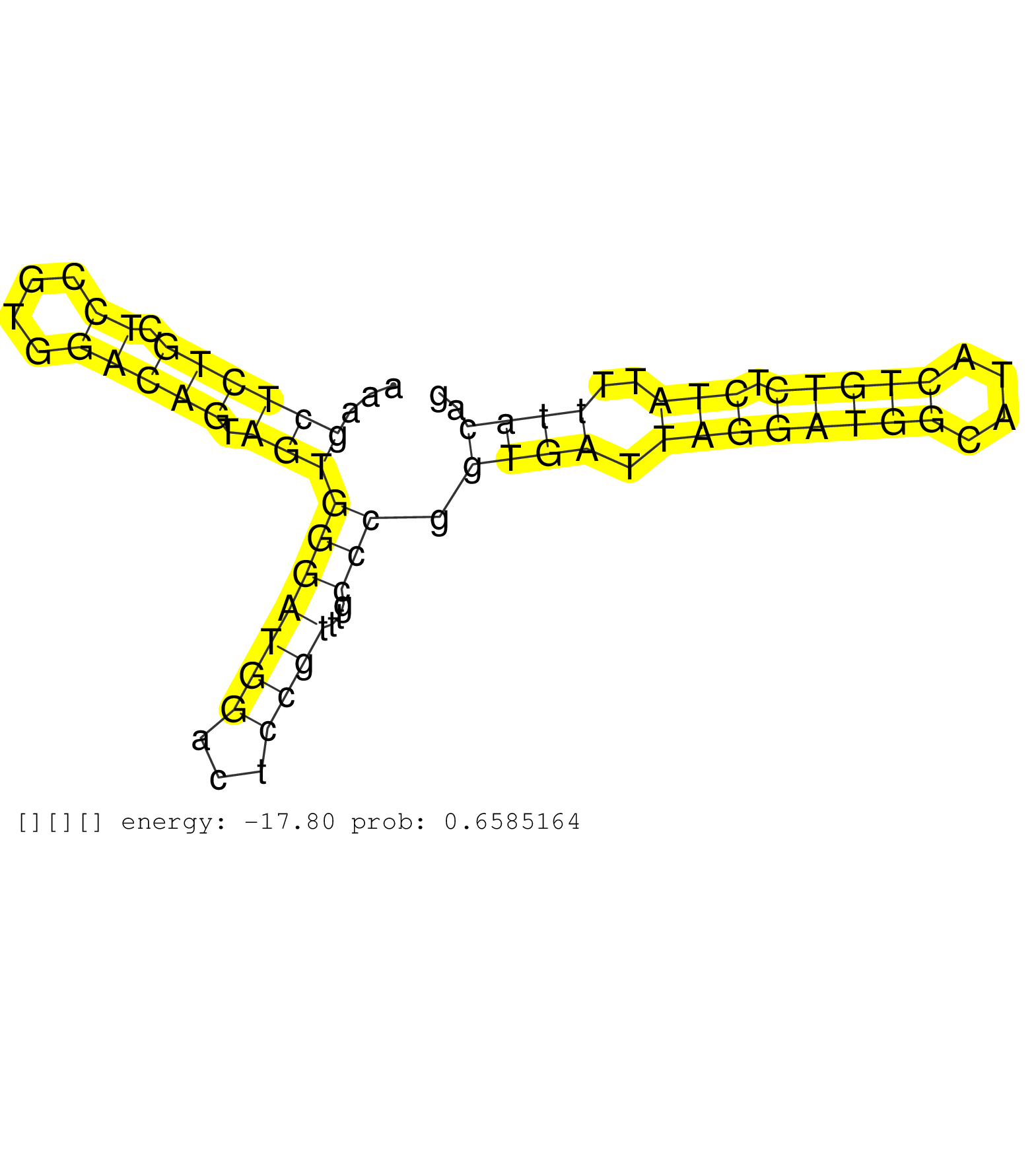

| Gene: Rccd1 | ID: uc009ial.1_intron_6_0_chr7_87466068_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| GCCAAAAGTAGAAGTGGTCAGCTGCAGACAGTTCAGCAGTCAAAAATCTGGTAGGACCTTTGGCCAAAGCTCTGCTCCGTGGACAGTAGTGGGATGGACTCCGTTTGCCCGGTGATTAGGATGGCATACTGTCTCTATTTTACAGGTAGTGCAACTTCAGCCCCCTGGAAGGCAGGATTTGCCAGCGTTCCTAAG ....................................................................((((((.((....))))).)))(((((((...))))...))).((((.((((((((....))))).)))..)))).................................................... .................................................................66.............................................................................145................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................TGATTAGGATGGCATACTGTCTCTATT........................................................ | 27 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TCTGGTAGGACCTTTGGC................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGATGGCATACTGTCTCTATTTTAC.................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................TGGTAGGACCTTTGGCCAAAGCTCTGC........................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................TCTGCTCCGTGGACAGTAGTGGGATGG.................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TCAAAAATCTGGTAGGACCTTTGGCCA................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCCGTGGACAGTAGTGGGATGGACTCCGT........................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGCCCGGTGATTAGGATGGCA..................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGCCCGGTGATTAGGATGGCATACTGTC.............................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GCTCCGTGGACAGTAGTGGGATGG.................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GTAGAAGTGGTCAGCTGCAGACAGTT.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................ACTCCGTTTGCCCGGTGATTAGGATG........................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....AAAGTAGAAGTGGTCAGCTGCAGACAGTT.................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................TGCAACTTCAGCCCCCTGGAAGGCAGGA.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................CAGTTCAGCAGTCAAAAATCTGGTAGt............................................................................................................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACCTTTGGCCAAAGCTC........................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..CAAAAGTAGAAGTGGTCAGCTGCAGAC...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGGCCAAAGCTCTGCTCCGTGGACAGTA........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGCTCCGTGGACAGTAGTGGGATGG.................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................TGGAAGGCAGGATTTGCCAGCGTTCC.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................TGCTCCGTGGACAGTAGTGGGATGGACT............................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CGGTGATTAGGATGGCATACTGTCTC............................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGATGGCATACTGTCTCTATTTTACAGc................................................. | 30 | c | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........GAAGTGGTCAGCTGCAGACAGT................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................TTGGCCAAAGCTCTGCTCCGTGGACAG............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TCTGCTCCGTGGACAGTAGTGGGATGGt................................................................................................. | 28 | t | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................TAGTGGGATGGACTCCGTTTGCCCGGT.................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GACAGTTCAGCAGTCAAAAATCTGGTA.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....AAAGTAGAAGTGGTCAGCTGCAGAC...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AAAGTAGAAGTGGTCAGCTGCAGACAGT................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........AGAAGTGGTCAGCTGCAGACAGTT.................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCCAAAAGTAGAAGTGGTCAGCTGCAGACAGTTCAGCAGTCAAAAATCTGGTAGGACCTTTGGCCAAAGCTCTGCTCCGTGGACAGTAGTGGGATGGACTCCGTTTGCCCGGTGATTAGGATGGCATACTGTCTCTATTTTACAGGTAGTGCAACTTCAGCCCCCTGGAAGGCAGGATTTGCCAGCGTTCCTAAG ....................................................................((((((.((....))))).)))(((((((...))))...))).((((.((((((((....))))).)))..)))).................................................... .................................................................66.............................................................................145................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................CTTTGGCCAAAGCTCTGCTCCGTGGACAa.............................................................................................................. | 29 | a | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TTTGGCCAAAGCTCTGCTCCGTGGACA.............................................................................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTCCGTTTGCCCGGTGAT............................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |