
| Gene: Zfp710 | ID: uc009hzi.1_intron_0_0_chr7_87169873_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(1) PIWI.mut |
(23) TESTES |
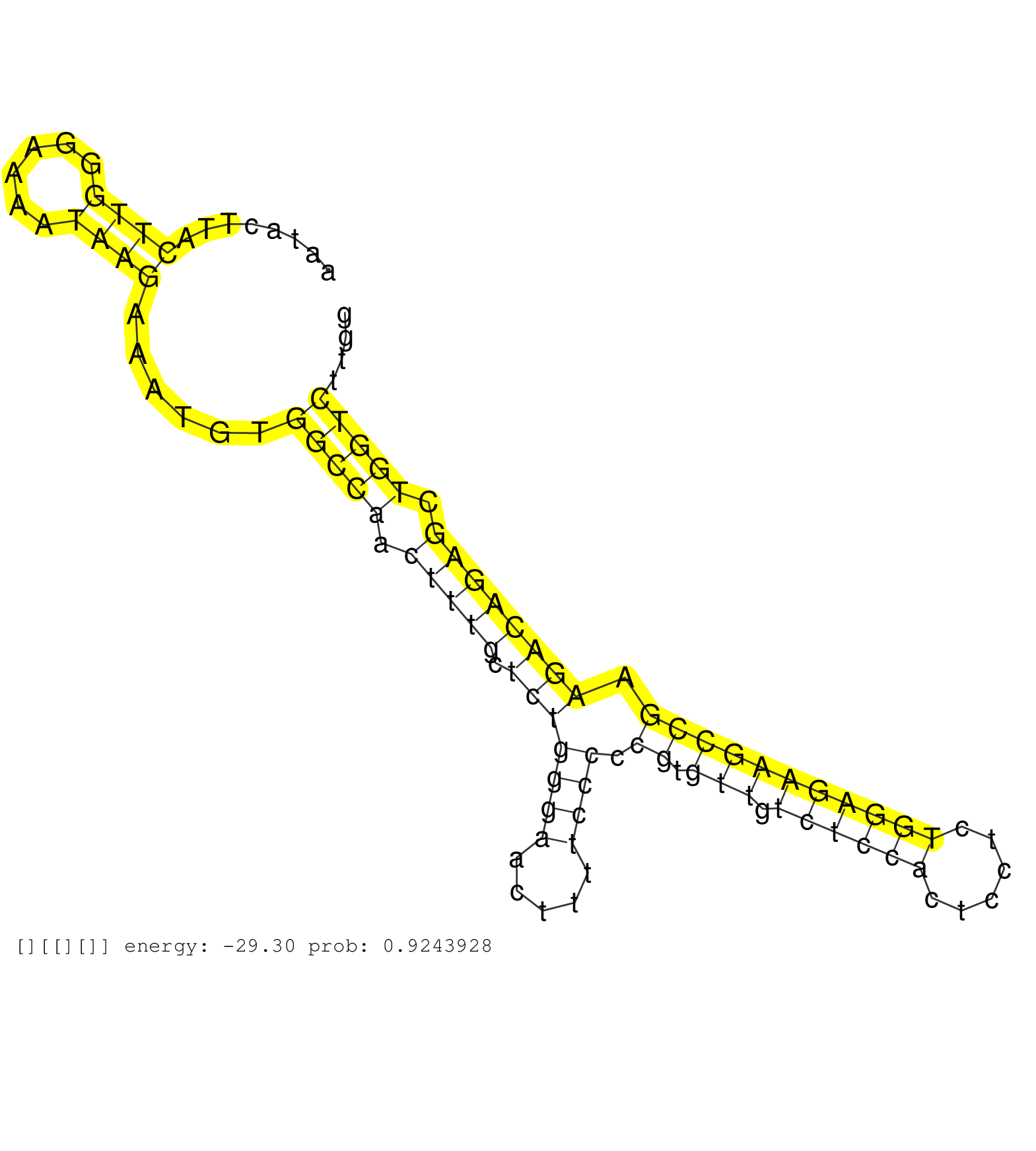
| ACGTTTCTCGCCCAGCTGCAGCCCCAGTCCAAGGGCCAGGCATCCAGCAGGTGCTCAATACTTACTTGGGAAAATAAGAAATGTGGCCAACTTTGCTCTGGGAACTTTTCCCCCGTGTTGTCTCCACTCCTCTGGAGAAGCCGAAGACAGAGCTGGTCTTGGGAGCACGCCCAGGCGCCCCATGAATAATTGACCCTTGGTGGCGACACTGCCCCGCTCTGTCCGCGGGCTCCGTACCCGCCCATGCGTC ................................................................((((......))))......(((((.(((((.(((((((.....)))).((.(((.((((((......))))))))))).)))))))).)))))............................................................................................ ........................................................57.......................................................................................................162...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................TGGAGAAGCCGAAGACAGAGCTGGTC............................................................................................ | 26 | 1 | 8.00 | 8.00 | - | 5.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................TCTGGAGAAGCCGAAGACAGAGCTGG.............................................................................................. | 26 | 1 | 6.00 | 6.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................TTACTTGGGAAAATAAGAAATGTGGCC.................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGGAGAAGCCGAAGACAGAGCTGGTCT........................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCTGGAGAAGCCGAAGACAGAGCTG............................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCTGGAGAAGCCGAAGACAGAGCTGGT............................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................................TGAATAATTGACCCTTGGTGGCGACACT........................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TACTTACTTGGGAAAATAAGAAATttg..................................................................................................................................................................... | 27 | ttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................CTCCTCTGGAGAAGCCGAAGACAGAGCT................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGAAATGTGGCCAACTTTGCTCTGGGA................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCGTGTTGTCTCCACTCCTCTGGAGAAG.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCTGGAGAAGCCGAAGACAGAGCTGGTC............................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCCTCTGGAGAAGCCGAAGACAGAGCT................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGAATAATTGACCCTTGGTGGCGACAC......................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTTCCCCCGTGTTGTCTCCACTCCT....................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCCCCCGTGTTGTCTCCACTCCTCTGat.................................................................................................................. | 28 | at | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGTCTCCACTCCTCTGGAGAAGCCG........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TACTTACTTGGGAAAATAAGAAATGTG..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................TACTTACTTGGGAAAATAAGAAATGcg..................................................................................................................................................................... | 27 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGGAGAAGCCGAAGACAGAGCTGG.............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................AATGTGGCCAACTTTGCTCTGGGAAC................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCAAGGGCCAGGCATCCAGCAGGTGC.................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCTGGAGAAGCCGAA......................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| ACGTTTCTCGCCCAGCTGCAGCCCCAGTCCAAGGGCCAGGCATCCAGCAGGTGCTCAATACTTACTTGGGAAAATAAGAAATGTGGCCAACTTTGCTCTGGGAACTTTTCCCCCGTGTTGTCTCCACTCCTCTGGAGAAGCCGAAGACAGAGCTGGTCTTGGGAGCACGCCCAGGCGCCCCATGAATAATTGACCCTTGGTGGCGACACTGCCCCGCTCTGTCCGCGGGCTCCGTACCCGCCCATGCGTC ................................................................((((......))))......(((((.(((((.(((((((.....)))).((.(((.((((((......))))))))))).)))))))).)))))............................................................................................ ........................................................57.......................................................................................................162...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................CTGCCCCGCTCTGTCCGC........................ | 18 | 1 | 6.00 | 6.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TCGCCCAGCTGCAGCCCCAGTCCAA.......................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AAGAAATGTGGCCAACTTTGCTCTGGGA................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................AGGTGCTCAATACTTACTTGG..................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GCCCCATGAATAATaatt............................................................ | 18 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................CCCAGGCGCCCCATGAAT............................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................CAGGCATCCAGCAGGTGCTCAATACT............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |