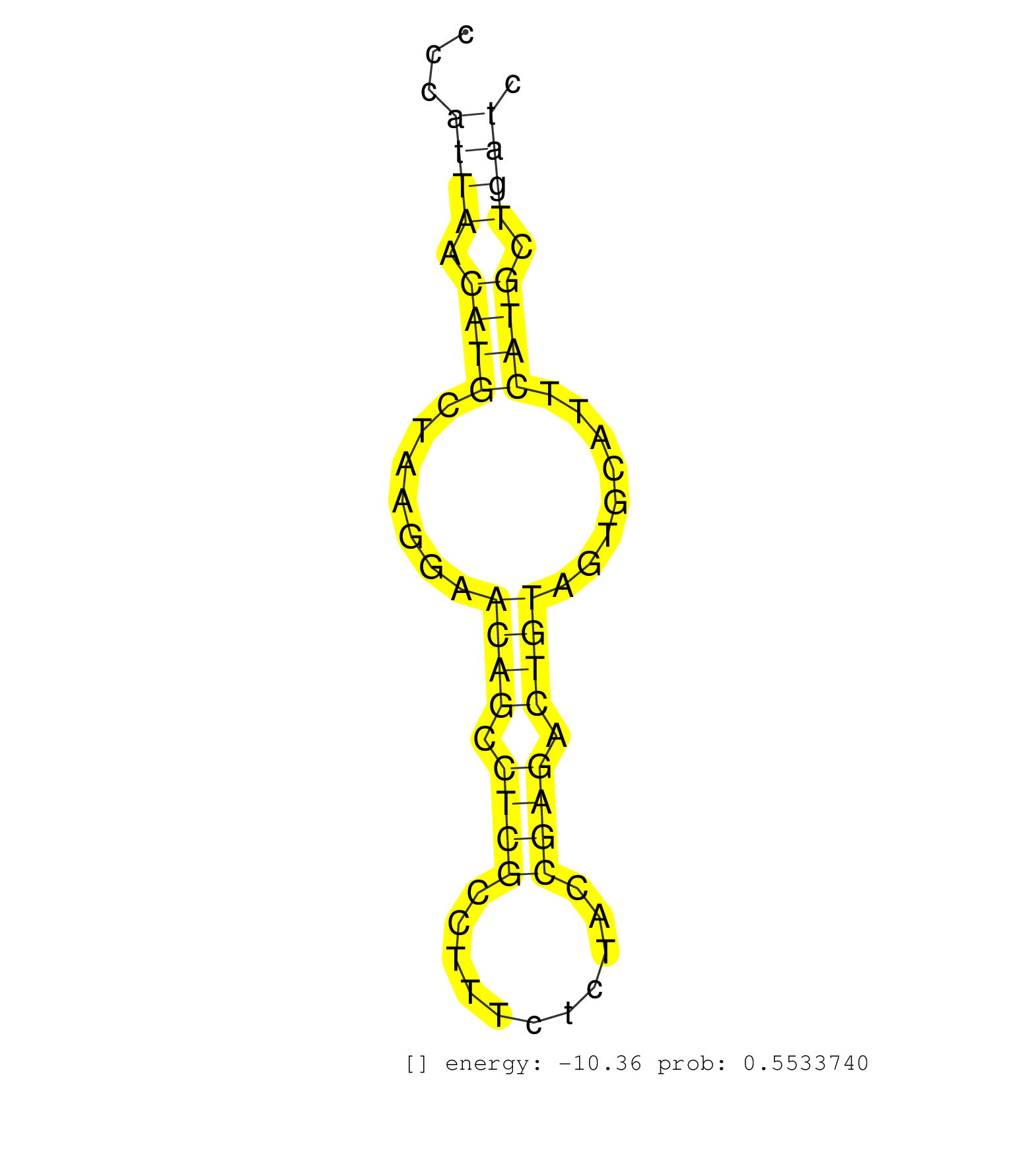

| Gene: Apba2 | ID: uc009hgj.1_intron_1_0_chr7_71698658_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| TTTCCTGCAGACAGGGACAGGGAGAGCCATCCTGTGTGTGGAGACTGCCGGTGAGTAGGATACATTTCCCATCTCCAGCTTTTCCTCGTGTACAATTGGGACAAAGCCCCCATTAACATGCTAAGGAACAGCCTCGCCTTTCTCTACCGAGACTGTAGTGCATTCATGCTGATCTTCAACTTTGTTGGGTTTGAAATATTCTCGTTTACAGTTGTCTCTTGGTATCCATGTGGGTTGGTTCCAAGGACCT ...............................................................................................................((((.((((.......((((.((((...........)))).))))........)))).))))............................................................................. ............................................................................................................109..............................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TACCGAGACTGTAGTGCATTCATGCT................................................................................ | 26 | 1 | 8.00 | 8.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................TACCGAGACTGTAGTGCATTCATGCTG............................................................................... | 27 | 1 | 7.00 | 7.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TACCGAGACTGTAGTGCATTCATGCTGATC............................................................................ | 30 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TAACATGCTAAGGAACAGCCTCGCCTTT............................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................GAGACTGCCGGTGAcaa................................................................................................................................................................................................. | 17 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................TACCGAGACTGTAGTGCATTCATGC................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................TGGAGACTGCCGGTGAGTAGGATAC........................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TTGTTGGGTTTGAAATATTCTCGTTT........................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGCAGACAGGGACAGGGAGAGCCATC........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................GAGACTGCCGGTGAcaaa................................................................................................................................................................................................ | 18 | caaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................TTCTCTACCGAGACTGTAGTGCATTCATGC................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TTCCTGCAGACAGGGACAGGGAGAGC............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACCGAGACTGTAGTGCATTCATGCTG............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................ACCGAGACTGTAGTGCATTCATGCTGATC............................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CGTTTACAGTTGTCTCTTGGTATCCATG.................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TATTCTCGTTTACAGTTGTCTCTTGG............................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGTGTGTGGAGACTGCCGGaaga................................................................................................................................................................................................... | 23 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................TACCGAGACTGTAGTGCATTCATGCTGA.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TTCTCTACCGAGACTGTAGTGCATTCATGCT................................................................................ | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCCGGTGAGTAGGATACATTTCCCATC................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................TGCTAAGGAACAGCCTCGCCTTTCTCTA........................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTTCCTGCAGACAGGGACAGGGAGAGCCATCCTGTGTGTGGAGACTGCCGGTGAGTAGGATACATTTCCCATCTCCAGCTTTTCCTCGTGTACAATTGGGACAAAGCCCCCATTAACATGCTAAGGAACAGCCTCGCCTTTCTCTACCGAGACTGTAGTGCATTCATGCTGATCTTCAACTTTGTTGGGTTTGAAATATTCTCGTTTACAGTTGTCTCTTGGTATCCATGTGGGTTGGTTCCAAGGACCT ...............................................................................................................((((.((((.......((((.((((...........)))).))))........)))).))))............................................................................. ............................................................................................................109..............................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................TCGTTTACAGTTtgcc..................................... | 16 | tgcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |