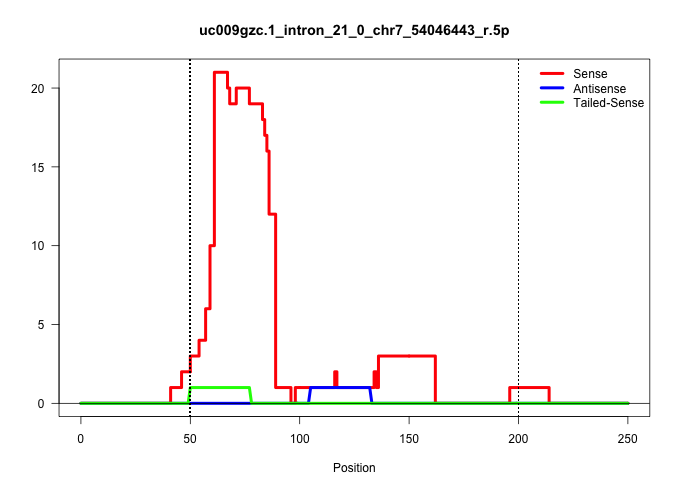
| Gene: Hps5 | ID: uc009gzc.1_intron_21_0_chr7_54046443_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| TCTGCTGCGCATCCTTCATCTCCAGCAGCGCCTCCAGGGCCTAGAACACGGTAGGCACTTAGAACGTTGAATGAATGAATGCATGCAAGCATGAGCGAGTCGTTAAGTTTTCTGAGGTAGAGTTTCCTGATCATTATTGGATAAGGAAGTTCCTATTGTGGAAATAAAGTCTATTAACTCCCCTTTGCATAATTTCTTCAATCGTTGTTACTTTGCGTCTTTCGAGCCTTTCCTTCCACGCACCTTGTGG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................GAACGTTGAATGAATGAATGCATGCAAG................................................................................................................................................................. | 28 | 1 | 11.00 | 11.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TAGAACGTTGAATGAATGAATGCATGC.................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................TTGGATAAGGAAGTTCCTATTGTGGA........................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| .......................................................................TGAATGAATGCATGCAAGCATGAGC.......................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CTTAGAACGTTGAATGAATGAATGCATGC.................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................TAGAACGTTGAATGAATGAATGCATG..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................TAGAACACGGTAGGCACTTAGAACGT....................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCACTTAGAACGTTGAATGAATGt............................................................................................................................................................................ | 28 | t | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................TATTGGATAAGGAAGTTCCTATTGTGGA........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................CACGGTAGGCACTTAGAACGTT...................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................GTAGAGTTTCCTGATCATT................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................GTCGTTAAGTTTTCTGAGG..................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TTCAATCGTTGTTACTTT.................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................CTTAGAACGTTGAATGAATGAATGCAT...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCACTTAGAACGTTGAATGAATG............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................GCACTTAGAACGTTGAATGAATGAATGCA....................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| TCTGCTGCGCATCCTTCATCTCCAGCAGCGCCTCCAGGGCCTAGAACACGGTAGGCACTTAGAACGTTGAATGAATGAATGCATGCAAGCATGAGCGAGTCGTTAAGTTTTCTGAGGTAGAGTTTCCTGATCATTATTGGATAAGGAAGTTCCTATTGTGGAAATAAAGTCTATTAACTCCCCTTTGCATAATTTCTTCAATCGTTGTTACTTTGCGTCTTTCGAGCCTTTCCTTCCACGCACCTTGTGG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................AGTTTTCTGAGGTAGAGTTTCCTGATCA..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |