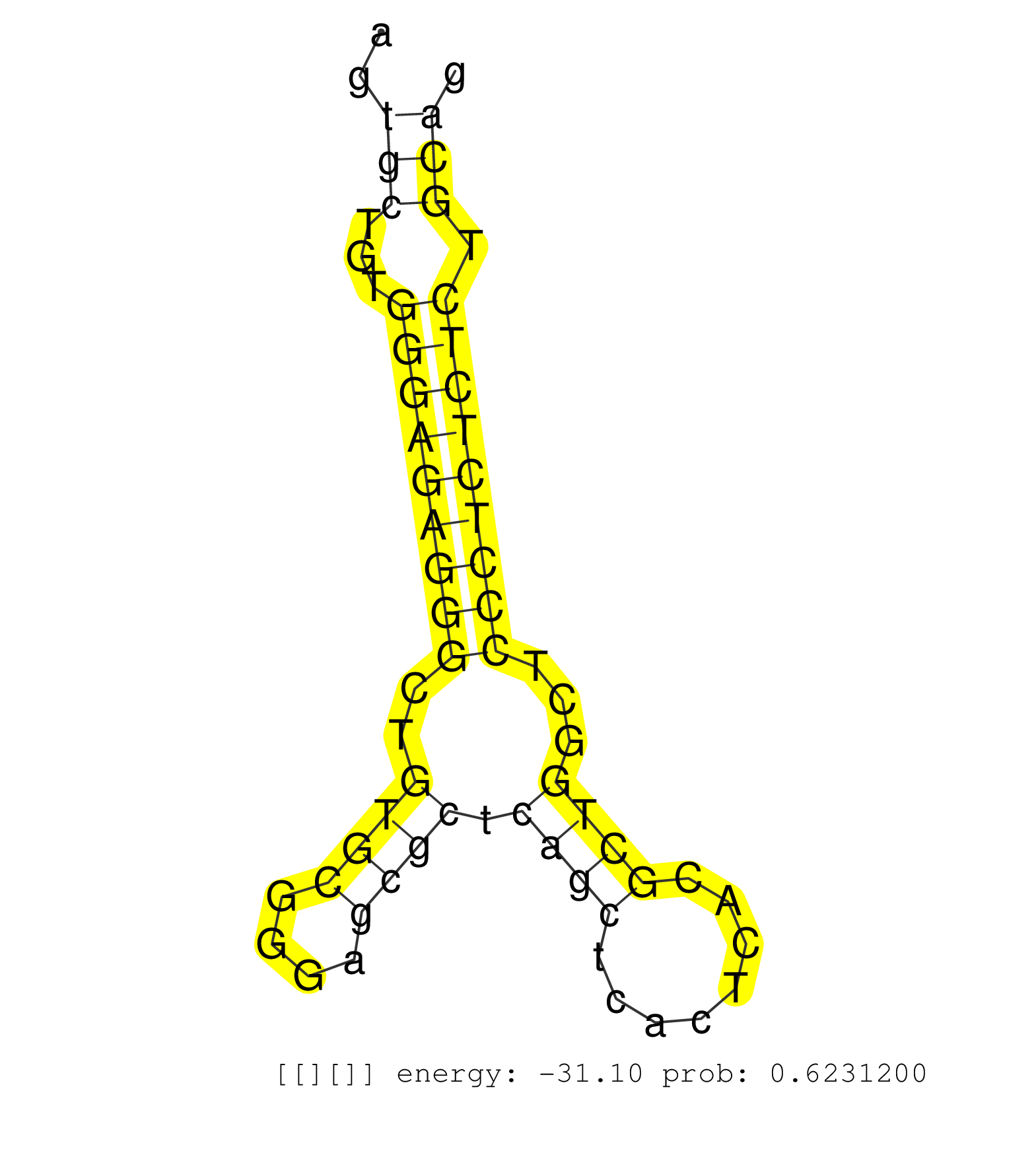

| Gene: Ruvbl2 | ID: uc009gvc.1_intron_0_0_chr7_52677376_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CACACAGTACATGAAGGAGTACCAAGATGCCTTCCTCTTCAATGAGCTCAGTGAGTGTCACCCTGGCTGCTAGTGCTGTGGGAGAGGGCTGTGCGGGAGCGCTCAGCTCACTCACGCTGGCTCCCTCTCTCTGCAGAAGGCGAAACAATGGACACCTCCTGAGCTGACCTGACACCCCAGCCCTTC .........................................................................(((...(((((((((..((((....)))).((((........))))...))))))))).)))................................................... .......................................................................72..............................................................136................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................CAATGGACACCTCCTGAGCTG.................... | 21 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCACGCTGGCTCCCTCTCTCTGC.................................................... | 23 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGTGGGAGAGGGCTGTGaa........................................................................................... | 19 | aa | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CTCTCTGCAGAAGGCGAAACAATGGACACC.............................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTCACCCTGGCTGCTAGTG............................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................TGTGGGAGAGGGCTGTGCGGt......................................................................................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCACGCTGGCTCCCTCTCTCTGCA................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...ACAGTACATGAAGGAGTACCAAGAT.............................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................GACCTGACACCCCAGCCC... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................TGGACACCTCCTGAGCTGACCTGACACC.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................ACACCTCCTGAGCTGACCTGACACCCCAGC..... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................TCACGCTGGCTCCCTCTCTCTGa.................................................... | 23 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CAATGGACACCTCCTGAGCTGACCTGAC............. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ACGCTGGCTCCCTCTCTCTGCAGt................................................. | 24 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTCACCCTGGCTGCTAGTGC.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................CTGTGGGAGAGGGCTGTGCGG.......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AATGGACACCTCCTGAGCTGACCTGACAC........... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| CACACAGTACATGAAGGAGTACCAAGATGCCTTCCTCTTCAATGAGCTCAGTGAGTGTCACCCTGGCTGCTAGTGCTGTGGGAGAGGGCTGTGCGGGAGCGCTCAGCTCACTCACGCTGGCTCCCTCTCTCTGCAGAAGGCGAAACAATGGACACCTCCTGAGCTGACCTGACACCCCAGCCCTTC .........................................................................(((...(((((((((..((((....)))).((((........))))...))))))))).)))................................................... .......................................................................72..............................................................136................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................CCTTCCTCTTCAATGAGCTCAGTGA.................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |