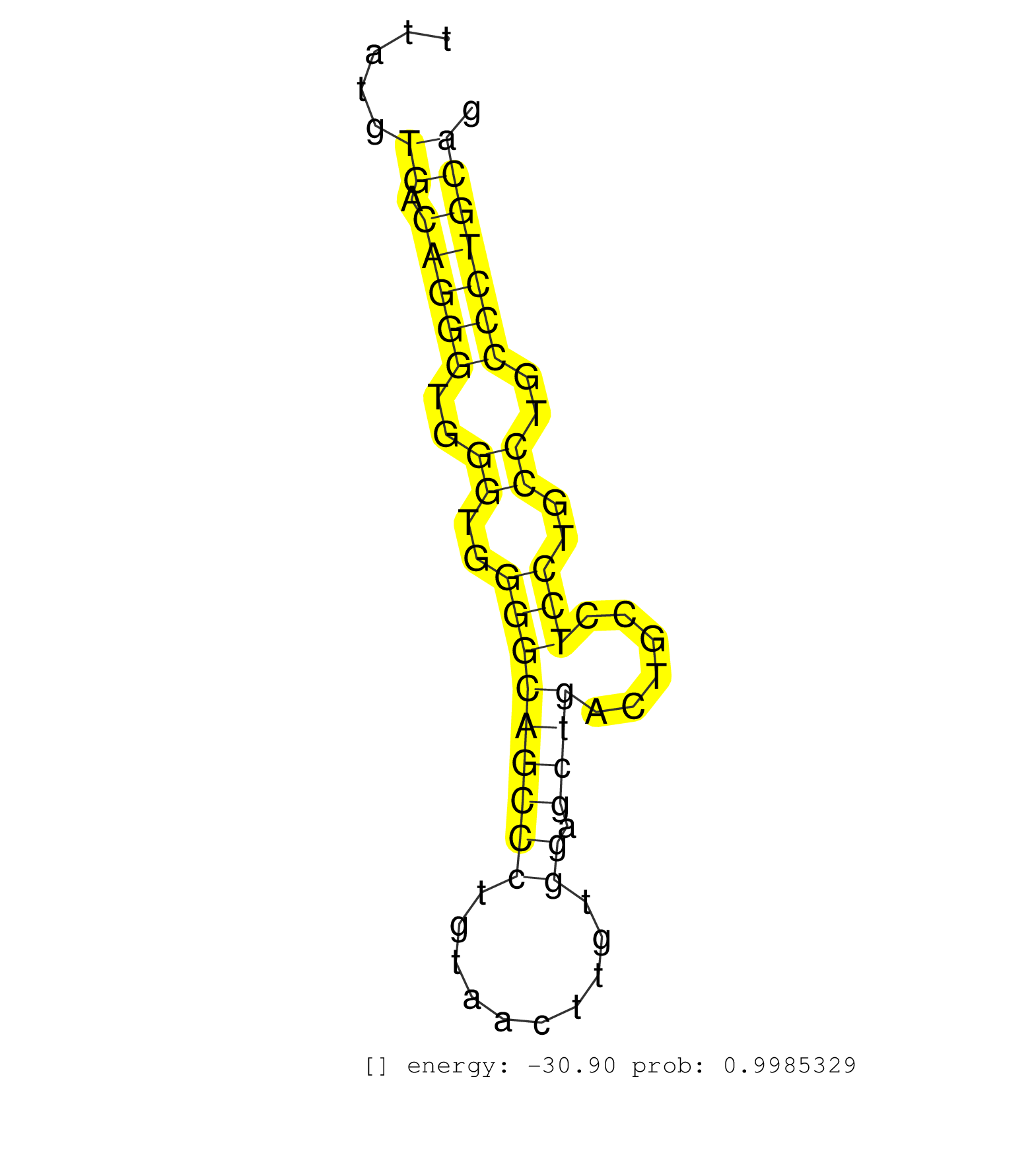
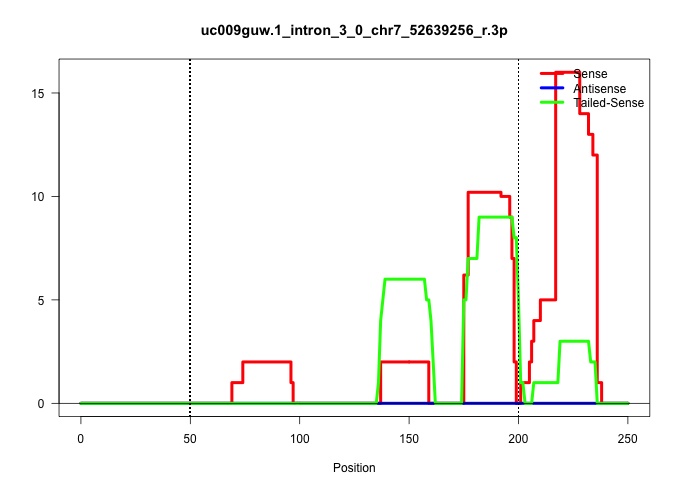
| Gene: Snrp70 | ID: uc009guw.1_intron_3_0_chr7_52639256_r.3p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(3) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| CCACATGTTGCCTCTGTGCCCCTCTGGTTTCCACTGGACTCTGACTTTTACTAGAGTGAGAGTGAGTCCCTTGGGACGTTTGAAGGATAACTTTGTTAATGGTGTCCCCCAGTCCAAGGCCTGTGTTATAAGTTATGTGACAGGGTGGGTGGGGCAGCCCTGTAACTTGTGGAGCTGACTGCCTCCTGCCTGCCCTGCAGATCCACATGGTATACAGTAAACGCTCTGGAAAACCCCGTGGTTATGCCTT .........................................................................................................................................((.(((((..((..(((((((((..........)).))))......)))..))..)))))))................................................... ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGC.................................................... | 23 | 1 | 33.00 | 33.00 | 5.00 | 11.00 | 2.00 | - | 4.00 | 8.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGC.................................................... | 21 | 1 | 12.00 | 12.00 | - | - | 8.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGCAGt................................................. | 26 | t | 8.00 | 0.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAGt................................................. | 24 | t | 7.00 | 0.00 | - | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTG..................................................... | 22 | 1 | 6.00 | 6.00 | - | - | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGCA................................................... | 24 | 1 | 6.00 | 6.00 | - | - | 1.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGtt................................................... | 24 | tt | 5.00 | 6.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGACAGGGTGGGTGGGGCAGC............................................................................................ | 21 | 1 | 5.00 | 5.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGCAt.................................................. | 25 | t | 4.00 | 6.00 | - | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAGA................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGa.................................................... | 23 | a | 3.00 | 6.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AACGCTCTGGAAAgtgc.............. | 17 | gtgc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAACTTGTGGAGCTGACTG..................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................AGCTGACTGCCTCCTGCCTGCCCTGCA................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAt.................................................. | 23 | t | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................CTCCTGCCTGCCCTGCAGt................................................. | 19 | t | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGACAGGGTGGGTGGGGCAGCC........................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ATGGTATACAGTAAACGCTCTG...................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ATGGTATACAGTAAACGCTCTGGAAA.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAta................................................. | 24 | ta | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGt.................................................... | 23 | t | 1.00 | 6.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTTGGGACGTTTGAAGGATAACTTTGT.......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACAGGGTGGGTGGGGCAGCCtat........................................................................................ | 23 | tat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................TATACAGTAAACGCTCTGGAAAACCCCG............ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................CTGCCTCCTGCCTGCCCTGCAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGTATACAGTAAACGCTCTGGAAtc................. | 26 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAGtat............................................... | 26 | tat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GTGACAGGGTGGGTGGGGCAGCCat......................................................................................... | 25 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAaa................................................. | 24 | aa | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TCCACATGGTATACAGTAAACGCTCTG...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAACTTGTGGAGCTGACTGC.................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGACAGGGTGGGTGGGGCAGCCaaa........................................................................................ | 25 | aaa | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGTATACAGTAAACGCTCTGGAAAAC................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGACAGGGTGGGTGGGGCAGt............................................................................................ | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGACAGGGTGGGTGGGGCAGCaa.......................................................................................... | 23 | aa | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................TGACAGGGTGGGTGGGGCAGCCa.......................................................................................... | 23 | a | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTG..................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GACAGGGTGGGTGGGGCAGCCCa......................................................................................... | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGatt.................................................. | 25 | att | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGTATACAGTAAACGCTCTG...................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGCAat................................................. | 26 | at | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACTGCCTCCTGCCTGCCCTGCAa.................................................. | 23 | a | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GACGTTTGAAGGATAACTTTGTT......................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCT...................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CATGGTATACAGTAAACGCTCTGGAAA.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTGCCCTGCAa.................................................. | 25 | a | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACAGGGTGGGTGGGGCAG............................................................................................. | 18 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACTGCCTCCTGCCTG.......................................................... | 17 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCACATGTTGCCTCTGTGCCCCTCTGGTTTCCACTGGACTCTGACTTTTACTAGAGTGAGAGTGAGTCCCTTGGGACGTTTGAAGGATAACTTTGTTAATGGTGTCCCCCAGTCCAAGGCCTGTGTTATAAGTTATGTGACAGGGTGGGTGGGGCAGCCCTGTAACTTGTGGAGCTGACTGCCTCCTGCCTGCCCTGCAGATCCACATGGTATACAGTAAACGCTCTGGAAAACCCCGTGGTTATGCCTT .........................................................................................................................................((.(((((..((..(((((((((..........)).))))......)))..))..)))))))................................................... ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|