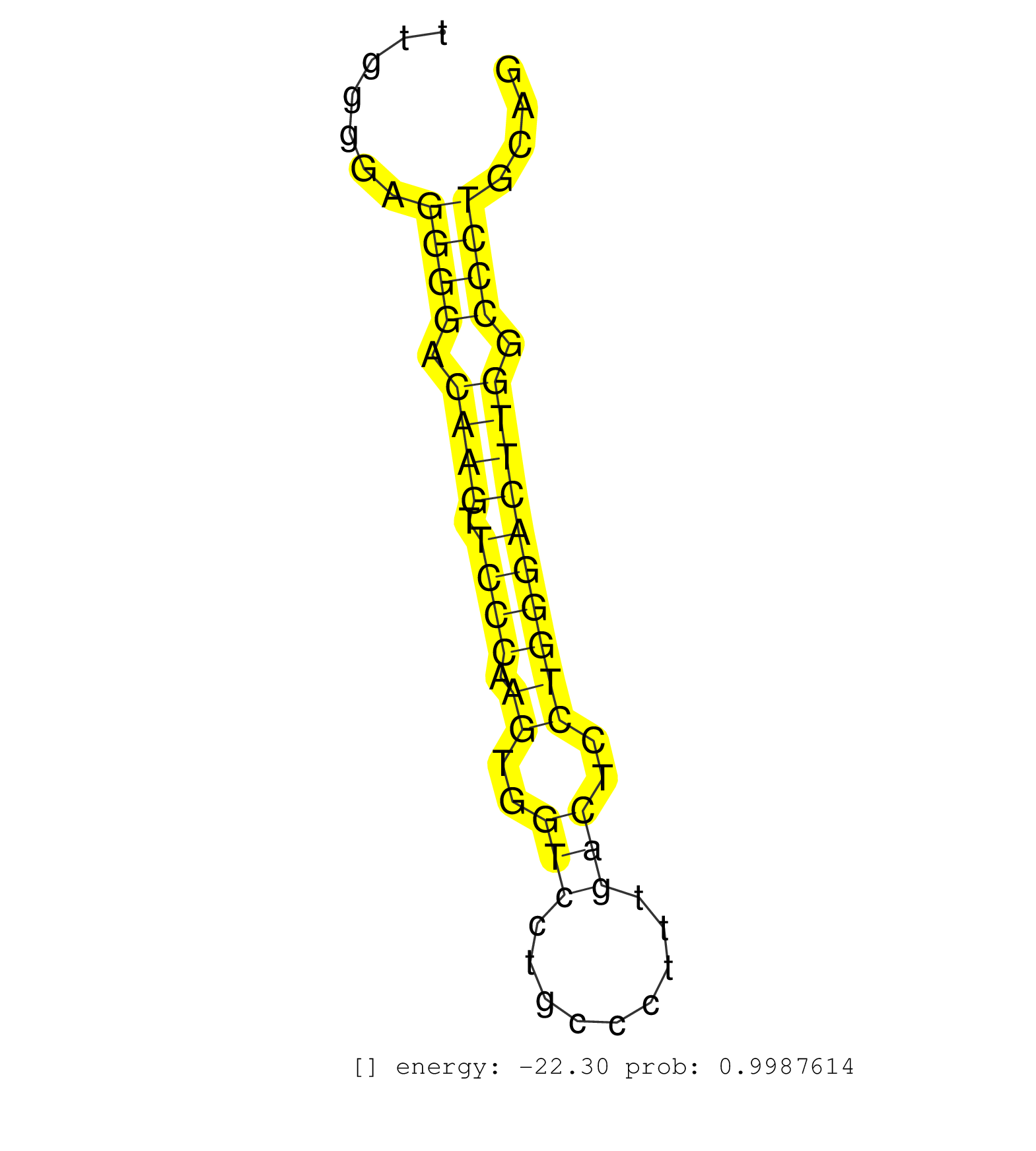
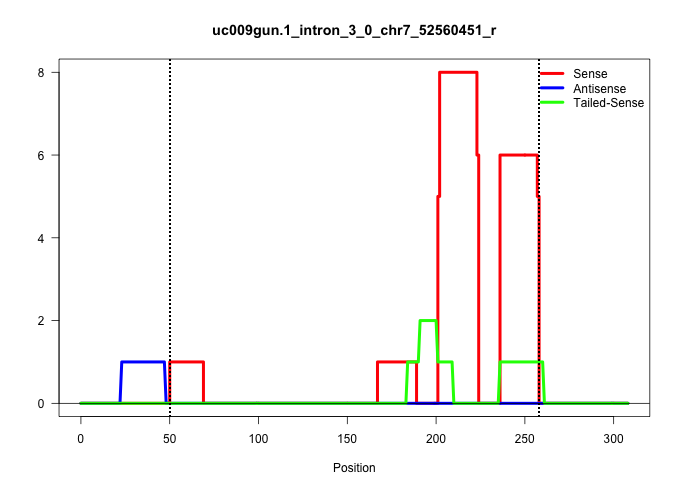
| Gene: Trpm4 | ID: uc009gun.1_intron_3_0_chr7_52560451_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) PIWI.mut |
(14) TESTES |
| CAGGTGTCGCAGGGCTAACTTGCCCGCCTCCCCAGTCTTCGAGCACTTCCGTGAGACTGGAGCTTGGTCTGCATGGGAGTGGGGCGGGGGGGGGGGGGCGTTTGGGGAGGGGAGAAGAAAAGCAGGTGGGTGGGGCTCGGGGAAGGTGTCAGGATCTGCACCCATCTGGGGACGAGGCATGAGGAATTTGGTGGCTTTGGGGAGGGGACAAGTTCCCAAGTGGTCCTGCCCTTTGACTCCTGGGACTTGGCCCTGCAGGTGTCTGTCTCTCTAAGGAAGCAGAGCGGAAGCTGCTGACTTGGGAGTCT ...........................................................................................................................................................................................................((((.((((.((((.((..(((.........)))..)))))))))).))))...................................................... ....................................................................................................................................................................................................197..........................................................258................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................CTCCTGGGACTTGGCCCTGCAG.................................................. | 22 | 1 | 8.00 | 8.00 | - | 3.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................GAGGGGACAAGTTCCCAAGTGGT.................................................................................... | 23 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................AGGGGACAAGTTCCCAAGTGGT.................................................................................... | 22 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AGGGGACAAGTTCCCAAGTGG..................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GGGGACGAGGCATGAGGAATTT....................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGGCTTTGGGGAGGGcgag.................................................................................................. | 19 | cgag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................................GGGGACAAGTTCCCAAGTGGTt................................................................................... | 22 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................AATTTGGTGGCTTgaga........................................................................................................... | 17 | gaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGACTGGAGCTTGGTC............................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................CTCCTGGGACTTGGCCCTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................CTCCTGGGACTTGGCCCTGCAGaaa............................................... | 25 | aaa | 1.00 | 8.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GAGGGGACAAGTTCCCAAGTGG..................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................CTCCTGGGACTTGGCCCTGCAGt................................................. | 23 | t | 1.00 | 8.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| CAGGTGTCGCAGGGCTAACTTGCCCGCCTCCCCAGTCTTCGAGCACTTCCGTGAGACTGGAGCTTGGTCTGCATGGGAGTGGGGCGGGGGGGGGGGGGCGTTTGGGGAGGGGAGAAGAAAAGCAGGTGGGTGGGGCTCGGGGAAGGTGTCAGGATCTGCACCCATCTGGGGACGAGGCATGAGGAATTTGGTGGCTTTGGGGAGGGGACAAGTTCCCAAGTGGTCCTGCCCTTTGACTCCTGGGACTTGGCCCTGCAGGTGTCTGTCTCTCTAAGGAAGCAGAGCGGAAGCTGCTGACTTGGGAGTCT ...........................................................................................................................................................................................................((((.((((.((((.((..(((.........)))..)))))))))).))))...................................................... ....................................................................................................................................................................................................197..........................................................258................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................AGCTGCTGACTTGGGAGgt... | 19 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................CCGCCTCCCCAGTCTTCGAGCACTT.................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ACTTGCCCGCCTCCCCAGTCTTCGA.......................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |