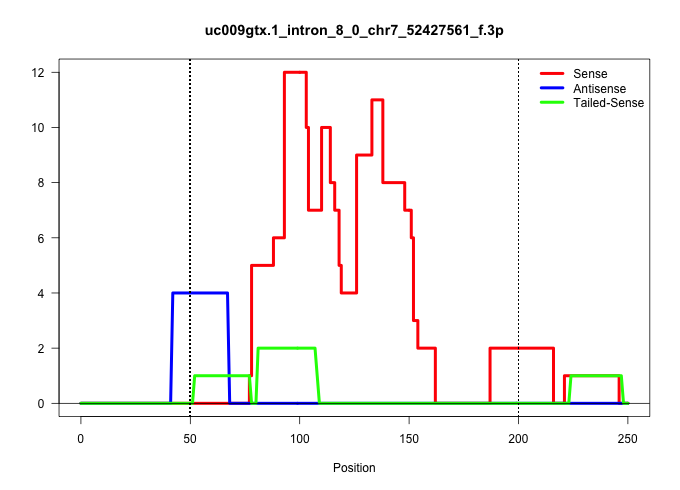
| Gene: Slc17a7 | ID: uc009gtx.1_intron_8_0_chr7_52427561_f.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| GAGCACCAAAGGGCATGAATCAGGTTGGGTCTTGGGGAAACAGGACCCAGTCTAGTTTGGTGGAACCAGGGTCAATTCTGCTGCAGGTCAGAATGAGTAGAAGGGGTCAGTAGAAGGGGGTCGGGGTGAGATCAAGACTGTCATTGGCGAGGTGGCCAACGGAGTCACAACGGGCACTTGGGAGACATGACCCTTTTCAGGTTTCGGGATGGAAGCCACGCTGCTGCTGGTGGTCGGATACTCGCACTCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TGAGATCAAGACTGTCATTGGCGAGG.................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAAGGGGGTCGGGGTGAGATCAAGAC................................................................................................................ | 28 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGCTGCAGGTCAGAATGAGTAGAAGG.................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TGACCCTTTTCAGGTTTCGGGATGGAAGC.................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AAGACTGTCATTGGCGAGGTGGCCAACGG........................................................................................ | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .............................................................................................TGAGTAGAAGGGGTCAGTAGAAGGG.................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................TGCAGGTCAGAATGAGTAGAAGGGcag.............................................................................................................................................. | 27 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................TAGTTTGGTGGAACCAGGGTCAATTt............................................................................................................................................................................ | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................TGAGTAGAAGGGGTCAGTAGAAGGGG................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGAGATCAAGACTGTCATTGGCGAG................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................TGAGTAGAAGGGGTCAGTAGAAG...................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................TGAGATCAAGACTGTCATTGGCGAGGTG................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CGGGGTGAGATCAAGACTGTCATTGGC...................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CAGAATGAGTAGAAGGGGTCAGTAGA........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGAGTAGAAGGGGTCAGTAGAAGGGGGT................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................CTGCTGCAGGTCAGAATGAGTAGAAG................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGCTGCTGGTGGTCGGATACTCGCA.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGCTGCAGGTCAGAATGAGTAGAAG................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................TGCAGGTCAGAATGAGTAGAAGGGGTCt............................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................TGAGTAGAAGGGGTCAGTAGA........................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGCTGGTGGTCGGATACTCGCcct.. | 24 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| GAGCACCAAAGGGCATGAATCAGGTTGGGTCTTGGGGAAACAGGACCCAGTCTAGTTTGGTGGAACCAGGGTCAATTCTGCTGCAGGTCAGAATGAGTAGAAGGGGTCAGTAGAAGGGGGTCGGGGTGAGATCAAGACTGTCATTGGCGAGGTGGCCAACGGAGTCACAACGGGCACTTGGGAGACATGACCCTTTTCAGGTTTCGGGATGGAAGCCACGCTGCTGCTGGTGGTCGGATACTCGCACTCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................GGACCCAGTCTAGTTTGGTGGAACCA...................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |