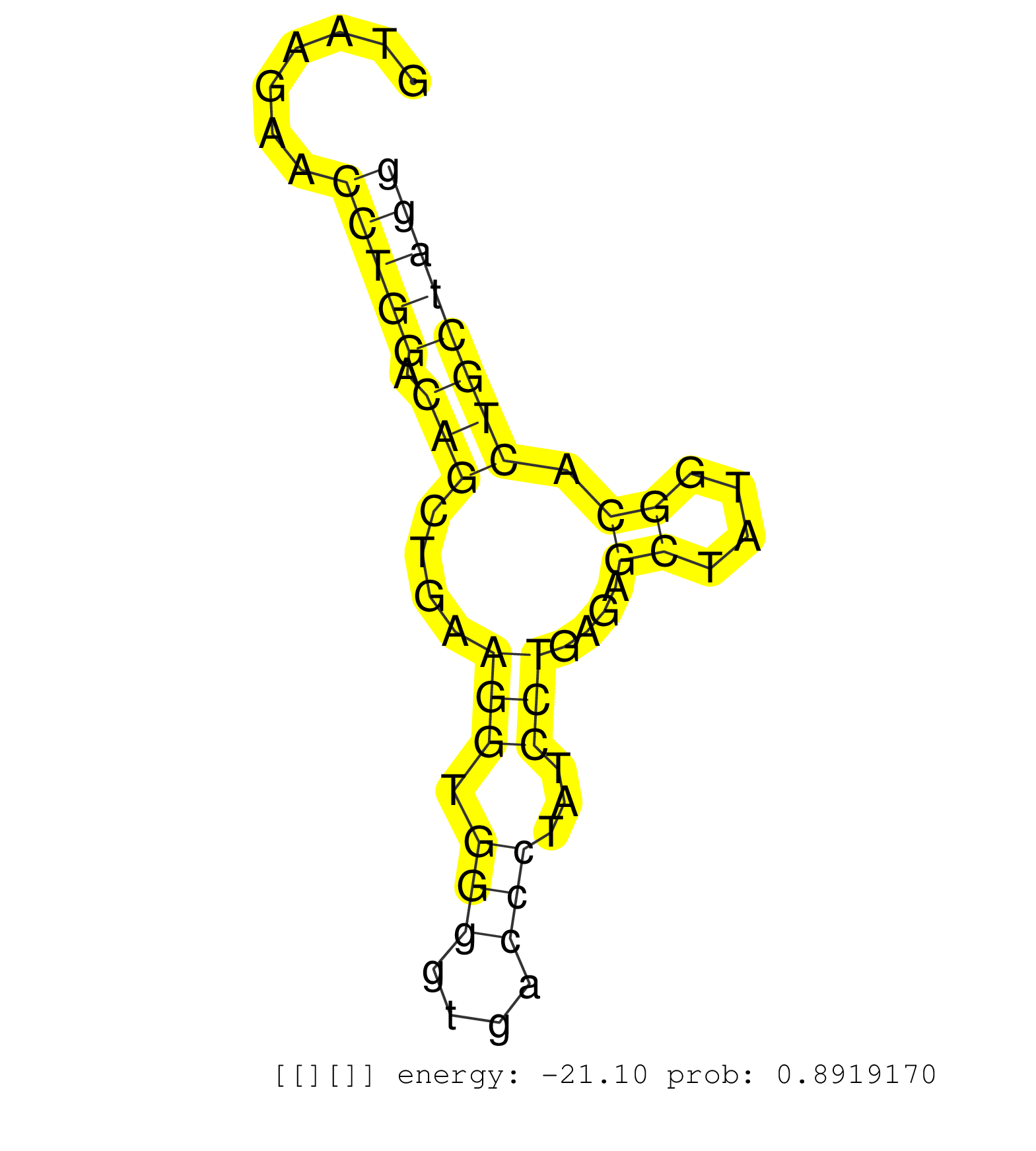
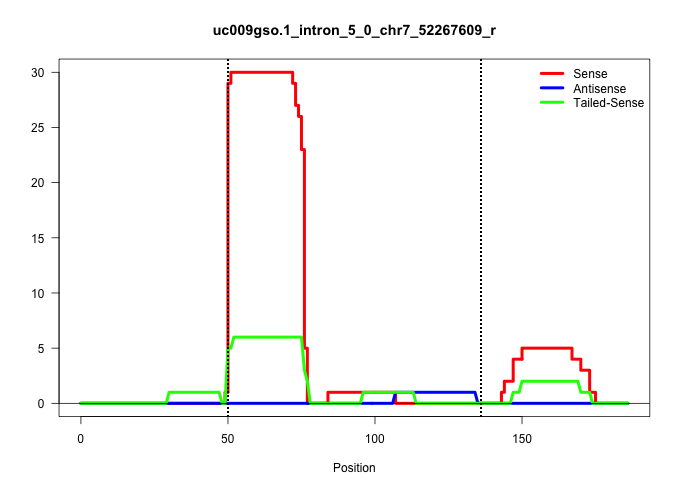
| Gene: Scaf1 | ID: uc009gso.1_intron_5_0_chr7_52267609_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| GATCCCCCGGACACCTGGGTTCCCAGCCGCCTGGACCTTCAGCCTGGCGAGTAAGAACCTGGACAGCTGAAGGTGGGGTGACCCTATCCTGAGAGCTATGGCACTGCTAGGGCTCACCATTTCCTCTGCCTTGCAGAAGTGAGGACATGCTAGAGCTGGTGGCTGAGGTCCGCATCGGTGACAGGG .........................................................(((((.(((....(((.(((....)))...)))....((....)).))))))))........................................................................... ..................................................51..........................................................111......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGAACCTGGACAGCTGAAGGTGG.............................................................................................................. | 26 | 1 | 17.00 | 17.00 | 2.00 | 3.00 | 6.00 | - | 2.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGGTGGG............................................................................................................. | 27 | 1 | 5.00 | 5.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................AGTGAGGACATGCTAGAGC.............................. | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................ATGCTAGAGCTGGTGGCTGAGGTCCGCA............ | 28 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGGTG............................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................GTGAGGACATGCTAGAGCTGGTGGCTGA.................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGGTGt.............................................................................................................. | 26 | t | 2.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCTAGAGCTGGTGGCTGAGGTCCGC............. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGGTGGtt............................................................................................................ | 28 | tt | 1.00 | 17.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGG................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TATCCTGAGAGCTATGGCACTGC............................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ACATGCTAGAGCTGGTGGCTGAGGTC................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGAACCTGGACAGCTGA.................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................AGTAAGAACCTGGACAGCTGAAGG................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGGTGttt............................................................................................................ | 28 | ttt | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCTAGAGCTGGTGGCTGAGGTCCGCc............ | 27 | c | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAG.................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TAAGAACCTGGACAGCTGAAGGTGG.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................CTGGACCTTCAGCCcctg.......................................................................................................................................... | 18 | cctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGGT................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................ACCTGGACAGCTGAAGGTGGGGT........................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................TAGAGCTGGTGGCTGAGGTCCGCAT........... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GACATGCTAGAGCTGGTGGCTGAG................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TATGGCACTGCTAGaatt........................................................................ | 18 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................AAGAACCTGGACAGCTGAAGGTGGt............................................................................................................. | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................TAGAGCTGGTGGCTGAGGTt................ | 20 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGAACCTGGACAGCTGAAGttgg.............................................................................................................. | 26 | ttgg | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GATCCCCCGGACACCTGGGTTCCCAGCCGCCTGGACCTTCAGCCTGGCGAGTAAGAACCTGGACAGCTGAAGGTGGGGTGACCCTATCCTGAGAGCTATGGCACTGCTAGGGCTCACCATTTCCTCTGCCTTGCAGAAGTGAGGACATGCTAGAGCTGGTGGCTGAGGTCCGCATCGGTGACAGGG .........................................................(((((.(((....(((.(((....)))...)))....((....)).))))))))........................................................................... ..................................................51..........................................................111......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TAGGGCTCACCATTTCCTCTGCCTTGCA................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CATCGGTGACAGagt.. | 15 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |