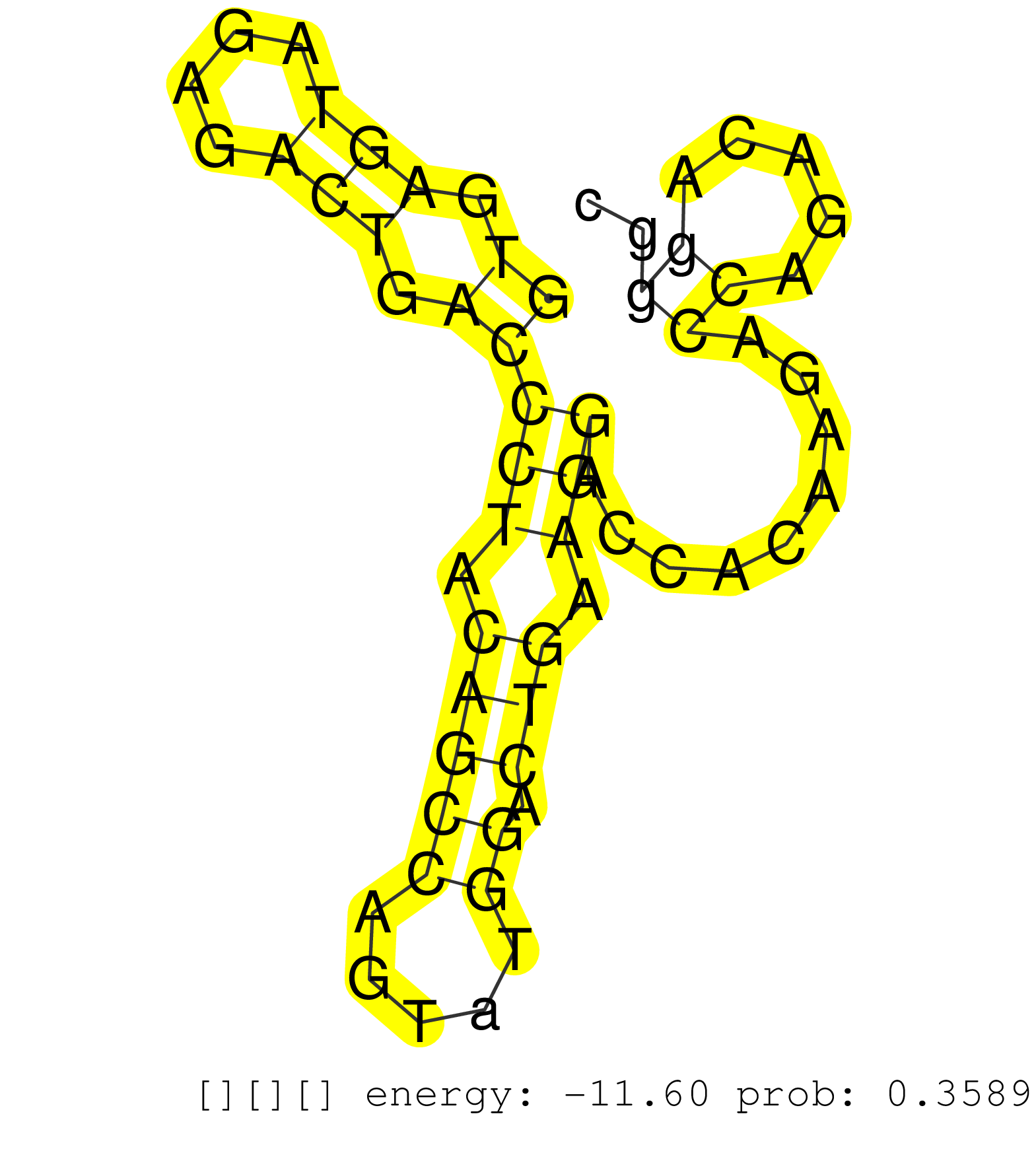

| Gene: Prmt1 | ID: uc009gsf.1_intron_7_0_chr7_52237264_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| CATCCAAAGACTACTACTTTGACTCCTATGCCCACTTTGGCATCCACGAGGTGAGTAGAGACTGACCCTACAGCCAGTATGGACTGAAGGACCACAAGACCAGACAGGGCTCTGAGTGTCTCAGCACGTTCACTGCTCCCTTTCTGCCCCTGGTCCACTTTTGAAGACTGAATTCTTCCATCTCTCCAGAAGCCAGTGCAAGTGCTTCCTGCCCCAGTGTACCACAGACCTAGGGATGAGGTCCGGCCCT ..................................................((.(((....))).))(((.(((((.....)).))).))).........((.....)).............................................................................................................................................. ..................................................51.........................................................110.......................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................TGGACTGAAGGACCACAAGACCAGACA................................................................................................................................................ | 27 | 1 | 7.00 | 7.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................TGAAGGACCACAAGACCAGACAGGGCTC.......................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAGAGACTGACCCTACAGCCAGT............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TCCAAAGACTACTACTTTGACTCCTATGC........................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........TACTACTTTGACTCCTATGCCCACTTT.................................................................................................................................................................................................................... | 27 | 2 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - |
| ...............................................................................TGGACTGAAGGACCACAAGACCAGACAGGGC............................................................................................................................................ | 31 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TACTACTTTGACTCCTATGCCCACTT..................................................................................................................................................................................................................... | 26 | 2 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 0.50 | - | 0.50 | - | - | - |
| ...............................................................................TGGACTGAAGGACCACAAGACCAGACAG............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....AAAGACTACTACTTTGAC................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................AAGGACCACAAGACCAGACAGGGCTCTGt....................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .ATCCAAAGACTACTACTTTGACTCCTA.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................TTTGAAGACTGAATTCTTCCATCTCTCC............................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................TGAAGGACCACAAGACCAGACAGGGCTCT......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................ACTGAAGGACCACAAGACCAGACAGGGCTCTGA....................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................CAGGGCTCTGAGTGTaagc............................................................................................................................... | 19 | aagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........TACTACTTTGACTCCTATGCCCcct...................................................................................................................................................................................................................... | 25 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGTAGAGACTGAaaaa..................................................................................................................................................................................... | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................TATGGACTGAAGGACCACAAGACCAGA.................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GACCACAAGACCAGACAGGGCTCTGAGTG.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGGACTGAAGGACCACAAGACCAGAC................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAGAGACTGACCCTACAGCC............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AAGACTACTACTTTGACTCCTATGC........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .ATCCAAAGACTACTACTTTGAC................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....AAAGACTACTACTTTGACTCCTATGCC.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................ACTGAAGGACCACAAGACCAGACAGGGCTC.......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TCCAAAGACTACTACTTTGA.................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CATCCAAAGACTACTACTTTGACTCCTATGCCCACTTTGGCATCCACGAGGTGAGTAGAGACTGACCCTACAGCCAGTATGGACTGAAGGACCACAAGACCAGACAGGGCTCTGAGTGTCTCAGCACGTTCACTGCTCCCTTTCTGCCCCTGGTCCACTTTTGAAGACTGAATTCTTCCATCTCTCCAGAAGCCAGTGCAAGTGCTTCCTGCCCCAGTGTACCACAGACCTAGGGATGAGGTCCGGCCCT ..................................................((.(((....))).))(((.(((((.....)).))).))).........((.....)).............................................................................................................................................. ..................................................51.........................................................110.......................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................GCCCCAGTGTACCACAGACCTA.................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCATCTCTCCAGAAGCCAGTGCAA................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCTGCCCCAGTGTACCACAGACCTA.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |