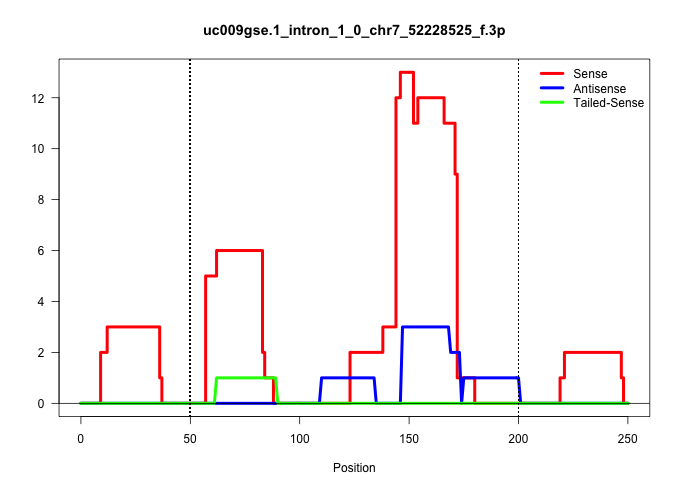
| Gene: AK020101 | ID: uc009gse.1_intron_1_0_chr7_52228525_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(16) TESTES |
| AGGAGACTATACTGAAGTTATTGGGGGAGGAGCTGCATGGGGAGGGCAGCAGGGGAGTGGAGTGAGTCTGGATGCTGAGCAGGGCAGGCCAGTGGGCAGCACAGAGCATCCCAGAGGAAGCCATAGTTAACCGTAGCCTGGGACTGGACATCCATATCTTCAGAATCGGTTTCAGGAAGCCCCGAAATCTGAATTCCCAGAAGGAAACCAGCCAATAGTTCTCCTCACCCAGACACGCCAGAGATGCCTT ....................................................((..((((......((((((((((.......((..(((....))).))....))))))).))).....))))......))....((((..(((((....))...((....))...)))..)))).......................................................................... ....................................................53.........................................................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TGGACATCCATATCTTCAGAATCGGTTT.............................................................................. | 28 | 1 | 7.00 | 7.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................TGGAGTGAGTCTGGATGCTGAGCAGG....................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TACTGAAGTTATTGGGGGAGGAGCTGC...................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ...........................................................................................................................TAGTTAACCGTAGCCTGGGACTGGACATC.................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGGACATCCATATCTTCAGAATCGGTT............................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TCCTCACCCAGACACGCCAGAGATGCC.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................TGGAGTGAGTCTGGATGCTGAGCAGGG...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................TGAGTCTGGATGCTGAGCAGGGCAGG.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............TGAAGTTATTGGGGGAGGAGCTGCA..................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTGAGTCTGGATGCTGAGCAGGG...................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCTCCTCACCCAGACACGCCAGAGATGC... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GACATCCATATCTTCAGAATCGGTTT.............................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................TGAGTCTGGATGCTGAGCAGGGCAGGaa................................................................................................................................................................ | 28 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TATCTTCAGAATCGGTTTCAGGAAGC...................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGGGACTGGACATCCATATCTTCAGAAT.................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGAGACTATACTGAAGTTATTGGGGGAGGAGCTGCATGGGGAGGGCAGCAGGGGAGTGGAGTGAGTCTGGATGCTGAGCAGGGCAGGCCAGTGGGCAGCACAGAGCATCCCAGAGGAAGCCATAGTTAACCGTAGCCTGGGACTGGACATCCATATCTTCAGAATCGGTTTCAGGAAGCCCCGAAATCTGAATTCCCAGAAGGAAACCAGCCAATAGTTCTCCTCACCCAGACACGCCAGAGATGCCTT ....................................................((..((((......((((((((((.......((..(((....))).))....))))))).))).....))))......))....((((..(((((....))...((....))...)))..)))).......................................................................... ....................................................53.........................................................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................CCAGAGGAAGCCATAGTTAACCGTA................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ACATCCATATCTTCAGAATCGGTTTCA............................................................................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ACATCCATATCTTCAGAATCGG................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GAAGCCCCGAAATCTGAATTCCCAGA................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |