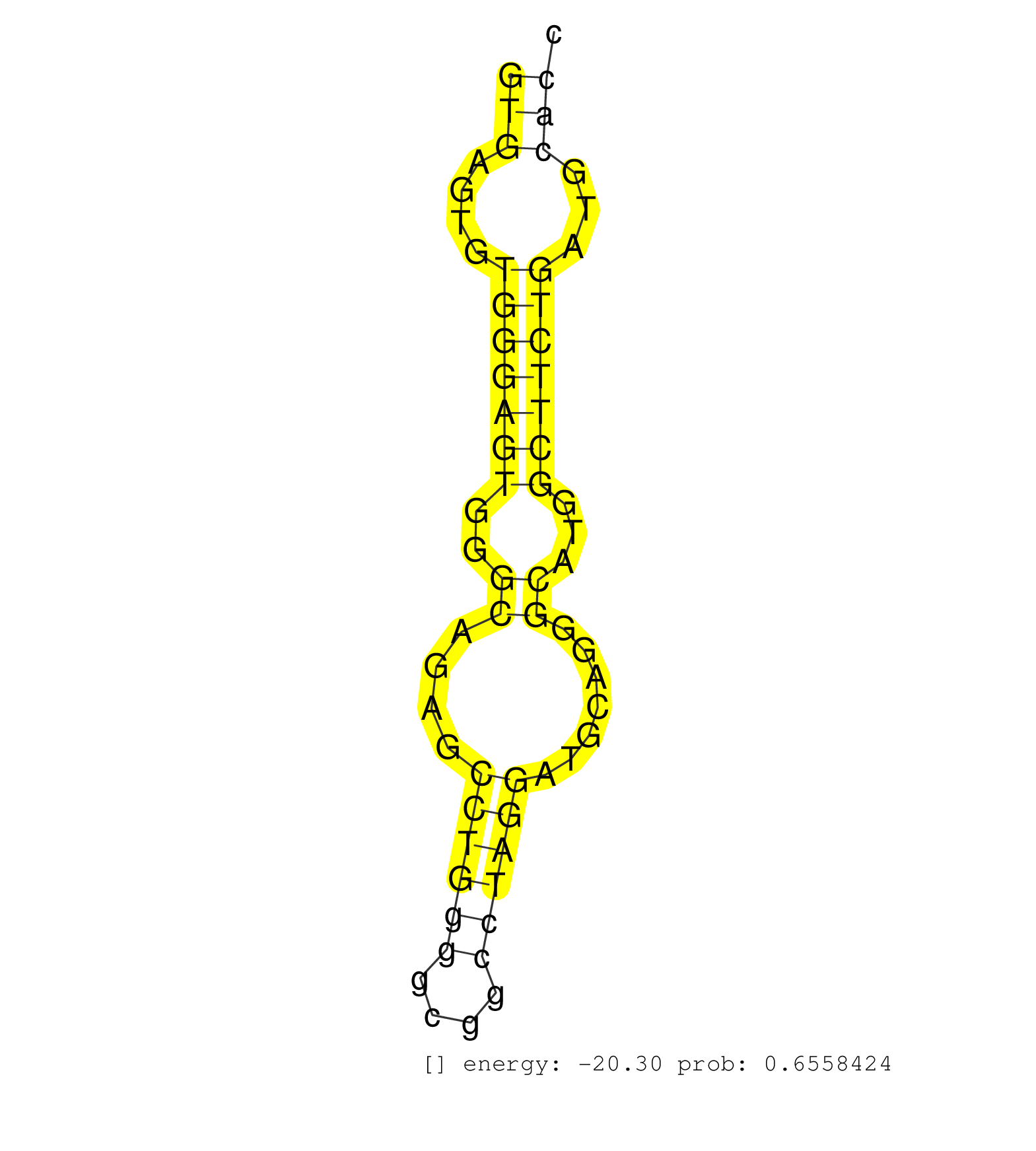

| Gene: Ptov1 | ID: uc009grk.1_intron_8_0_chr7_52122488_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(22) TESTES |
| CTGAAACGCACCCTGCCCTGCCAGGCCTATGTGAACCAGGGCGAGAACCTGTGAGTGTGGGAGTGGGCAGAGCCTGGGGCGGCCTAGGATGCAGGGCATGGCTTCTGATGCACCCCCTACTGCAGGGAGACGGACCAGTGGCCCCAGAAGTTGATCATGCAGCTCATCCCACAGC ..................................................(((....(((((((..((....((((((....)))))).......))...)))))))...))).............................................................. ..................................................51.............................................................114........................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTGG.................................................................................................. | 27 | 1 | 11.00 | 11.00 | 1.00 | 3.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTG................................................................................................... | 26 | 1 | 10.00 | 10.00 | 1.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..................TGCCAGGCCTATGTGAACCAGGGCGAG.................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | - | - | 2.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AACCAGGGCGAGAACCT............................................................................................................................. | 17 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTGt.................................................................................................. | 27 | t | 3.00 | 10.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TGAGTGTGGGAGTGGGCAGAGCCTGG.................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAG....................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................CGGCCTAGGATGCAGGG............................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGCCAGGCCTATGTGAACCAGGGC..................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TAGGATGCAGGGCATGGCTTCTGATG................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGGCCCCAGAAGTTGATCATGCAGCTC.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGTGGGAGTGGGCAGAGCCTGGGGCGGCC........................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TATGTGAACCAGGGCGAG.................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TATGTGAACCAGGGCGAGAACCTGgag......................................................................................................................... | 27 | gag | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTGaa................................................................................................. | 28 | aa | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTGtt................................................................................................. | 28 | tt | 1.00 | 10.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGC...................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TACTGCAGGGAGACGGACCAGTGG.................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................CTGCCAGGCCTATGTGAAC........................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GCCAGGCCTATGTGAACCAGGGCGAG.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................GACGGACCAGTGGCCCCAGAAGTTGATC................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGTGAACCAGGGCGAGA................................................................................................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCAGGGAGACGGACCAGTGG.................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....AACGCACCCTGCCCTGCCAGGCC.................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................AGTTGATCATGCAGCTCATCCCA.... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................CGGCCTAGGATGCAGGGCATGG.......................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCT.................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTGAGTGTGGGAGTGGGCAGAGCaat................................................................................................... | 27 | aat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................GAGTGGGCAGAGCCTGGGGCGGCCTA......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGCCAGGCCTATGTGAACCAGGGCGgg.................................................................................................................................. | 27 | gg | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................TAGGATGCAGGGCATGGCTTCTG.................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GGAGACGGACCAGTGGCCCCAGAA.......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTGGG................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CGGACCAGTGGCCCCAGAAGTTGATCAT................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................TGATGCACCCCCTACTGCAGGGAGAC............................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGCCAGGCCTATGTGAACCAGGGCG.................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTAGGATGCAGGGCATGGCT........................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................GTGAACCAGGGCGAGAAatt............................................................................................................................. | 20 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGAGTGGGCAGAGCCTata................................................................................................. | 28 | ata | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................ATGTGAACCAGGGCGAGA................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGAAACGCACCCTGCCCTGCCAGGCCTATGTGAACCAGGGCGAGAACCTGTGAGTGTGGGAGTGGGCAGAGCCTGGGGCGGCCTAGGATGCAGGGCATGGCTTCTGATGCACCCCCTACTGCAGGGAGACGGACCAGTGGCCCCAGAAGTTGATCATGCAGCTCATCCCACAGC ..................................................(((....(((((((..((....((((((....)))))).......))...)))))))...))).............................................................. ..................................................51.............................................................114........................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|