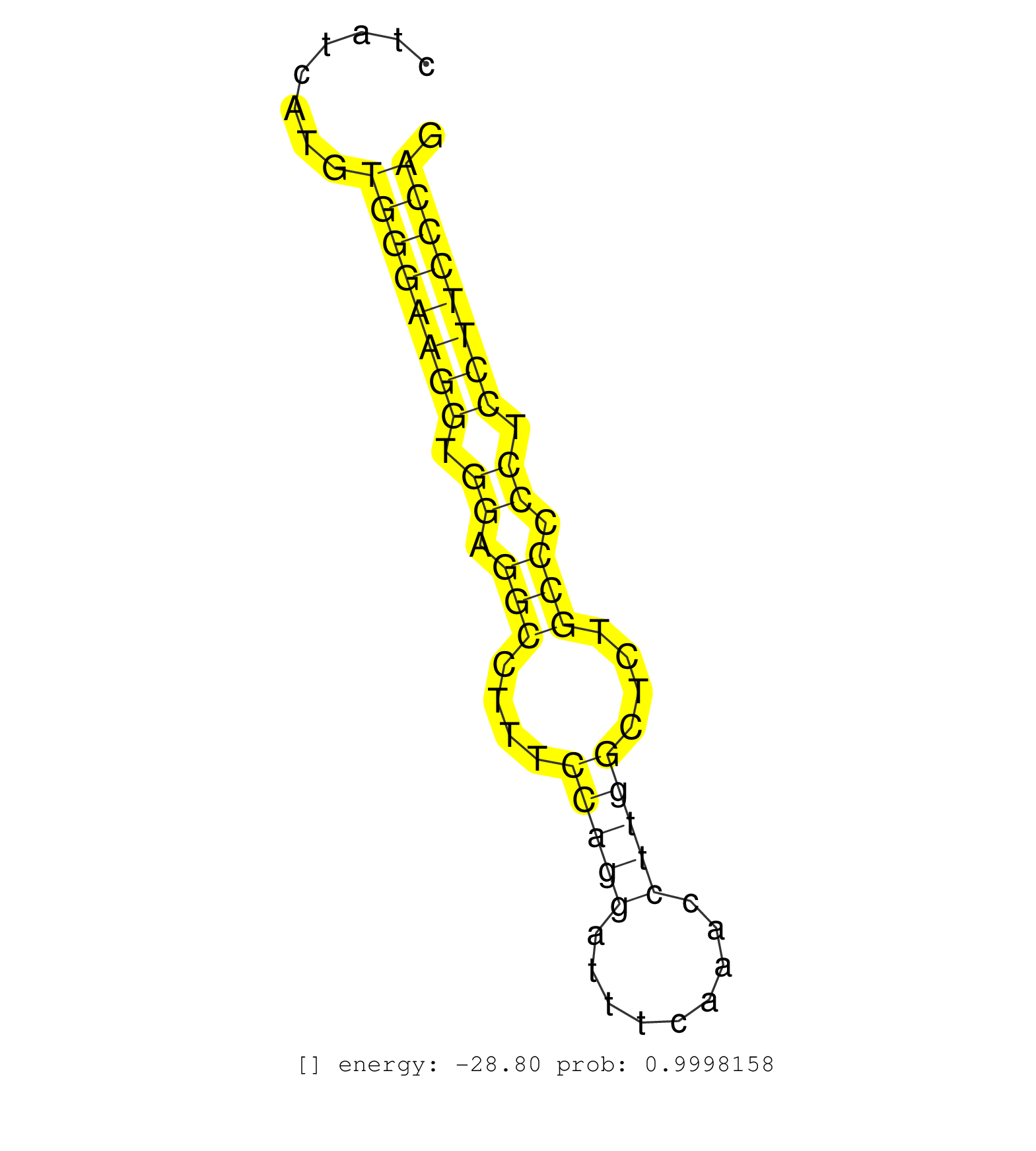
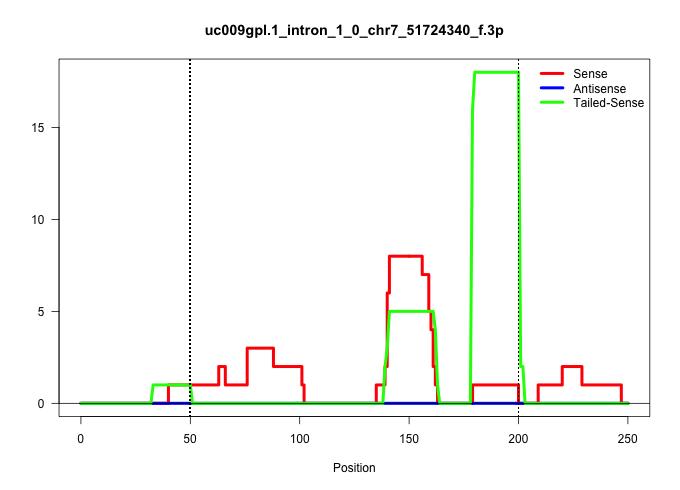
| Gene: Josd2 | ID: uc009gpl.1_intron_1_0_chr7_51724340_f.3p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(3) PIWI.ip |
(20) TESTES |
| TGAGTGGGAAAAGATAAAGCCTTAGCCAGTCTAAGAGTACAGGGCTCAGGGGTGGAGGGAGAACAGGACGTGAGGATGATAAACTGTGTGGGGGTTGAAGAGAGACGCGTCAAAGGCCAGGCTCTGGGCCTCTCCTATCATGTGGGAAGGTGGAGGCCTTTCCAGGATTTCAAACCTTGGCTCTGCCCCCTCCTTCCCAGGCTAGCCCCAGATTCCCGGCTGAACCCCCATCGCAGTCTCTTGGGCACCG ..............................................................................................................................................((((((((.((.(((....(((((.........)))))....))).)).))))))))................................................... ......................................................................................................................................135..............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................GCTCTGCCCCCTCCTTCCCAGt................................................. | 22 | t | 21.00 | 1.00 | 6.00 | 5.00 | - | 1.00 | 1.00 | 3.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................CTGTGTGGGGGTTGAAGA..................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ATGTGGGAAGGTGGAGGCCTTTCt....................................................................................... | 24 | t | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAAGGTGGAGGCCTTT......................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GCTCTGCCCCCTCCTTCCCcgt................................................. | 22 | cgt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GCTCTGCCCCCTCCTTCCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................GTGGAGGGAGAACAGGACGTGAGGATGATA......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGATAAACTGTGTGGGGGTTGAAGA..................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CTCTGCCCCCTCCTTCCCAGttt............................................... | 23 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................CAGGACGTGAGGATGATAAACTGTG.................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................TGTGGGAAGGTGGAGGCCTTTt........................................................................................ | 22 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAAGGTGGAGGCCT........................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGGGAAGGTGGAGGCCTTTCaa...................................................................................... | 23 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGGGAAGGTGGAGGCCTTTCC....................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGAGTACAGGGCTCctaa....................................................................................................................................................................................................... | 18 | ctaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGGGAAGGTGGAGGCCTTTCa....................................................................................... | 22 | a | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GCTCTGCCCCCTCCTTCCCAGa................................................. | 22 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GCTCTGCCCCCTCCTTCCCA................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AGATTCCCGGCTGAACCCCC..................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................TGAACCCCCATCGCAGTCTCTTGGGCA... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGGGAAGGTGGAGGCCTTTC........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAAGGTGGAGGCCTT.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................ATGTGGGAAGGTGGAGGCCT........................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTGAGGATGATAAACTGTGTGGGGG............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CTCTGCCCCCTCCTTCCCAtttt............................................... | 23 | tttt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TATCATGTGGGAAGGTGGAGG.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................AGGGCTCAGGGGTGGAGGGAGAACAG........................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................TGATAAACTGTGTGGGGGTTGAAGAG.................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| TGAGTGGGAAAAGATAAAGCCTTAGCCAGTCTAAGAGTACAGGGCTCAGGGGTGGAGGGAGAACAGGACGTGAGGATGATAAACTGTGTGGGGGTTGAAGAGAGACGCGTCAAAGGCCAGGCTCTGGGCCTCTCCTATCATGTGGGAAGGTGGAGGCCTTTCCAGGATTTCAAACCTTGGCTCTGCCCCCTCCTTCCCAGGCTAGCCCCAGATTCCCGGCTGAACCCCCATCGCAGTCTCTTGGGCACCG ..............................................................................................................................................((((((((.((.(((....(((((.........)))))....))).)).))))))))................................................... ......................................................................................................................................135..............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|