
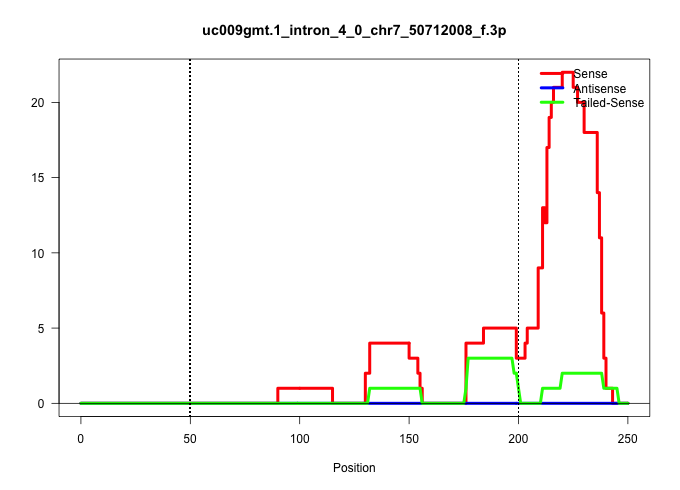
| Gene: Etfb | ID: uc009gmt.1_intron_4_0_chr7_50712008_f.3p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| GTAAAGCCTCCTAGGGCATCCTAGCCTGAATGGATAGACTACATCAGGTTTAGCTATGAGAAATCACATTCCAGGCCAGCAGAGAGGTCCTGAGGGATTCTGGCAACAGATATGGCTTTAAACATGCACTACTCACCAGGAGGAAGTTGGGTCAGGTAAGGGCTTCGCAGCCCCCCTGAGCCACTGATTCCTGTGTGCAGAAAGCCAAGAAGAAGAAGATTGAAGTGGTCAAGGCTGGAGACCTGGGCGT ...................................................................................................................................(((((((((((..((.(((.(((((...(((((....))))).))))).)))))..)))))).))).)).................................................. ...............................................................................................................................128.....................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................GAAGAAGATTGAAGTGGTCAAGGCTG............. | 26 | 1 | 12.00 | 12.00 | 10.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AAGAAGAAGATTGAAGTGGTCAAGGCT.............. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGAAGATTGAAGTGGTCAAGGCTGG............ | 25 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCACCAGGAGGAAGTTGGGTCAGa.............................................................................................. | 24 | a | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................GAAGAAGATTGAAGTGGTCAAGGCTGG............ | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCACTGATTCCTGTGTGCA................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTCACCAGGAGGAAGTTGGGTCA................................................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GAGCCACTGATTCCTGTGTGCAGt................................................. | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................ACTCACCAGGAGGAAGTTGGGTCAGG.............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................................TGATTCCTGTGTGCAGAAAGCCAAGAAG...................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCACCAGGAGGAAGTTGGGTCAG............................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAAAGCCAAGAAGAAGAAGATTGAAG......................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCACTGATTCCTGTGTG..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................CCAAGAAGAAGAAGATTGAAGTGGTC.................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AGATTGAAGTGGTCAAGGCTGGAGACC....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................GCCAAGAAGAAGAAGATTGAAGTGGTC.................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AAGATTGAAGTGGTCAAGGCTGGAG.......... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGAAGATTGAAGTGGTCAAGGCT.............. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GAAGATTGAAGTGGTCAAcg................ | 20 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GAAGATTGAAGTGGTCAAGGCTGGAG.......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................................................TGAAGTGGTCAAGGCTGGA........... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCACTGATTCCTGTGTGCAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GAGCCACTGATTCCTGTGTGt.................................................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GAAGAAGATTGAAGTGGTCAAGGCTGGt........... | 28 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCACTGATTCCTGTGTGCAt.................................................. | 24 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AAAGCCAAGAAGAAGAAGATTGAAGTG....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AAGAAGAAGATTGAAGTGGTCAAGGCTG............. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCACCAGGAGGAAGTTGGGTCA................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGAAGATTGAAGTGGTCAAGGCTGGA........... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGAAGTGGTCAAGGCTGGAGACCTGa.... | 26 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GAAGATTGAAGTGGTCAAGGCTGGA........... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................ACTCACCAGGAGGAAGTTGG.................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AGCCACTGATTCCTGTGTGCAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................TGAGGGATTCTGGCAACAGATATGG....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GTAAAGCCTCCTAGGGCATCCTAGCCTGAATGGATAGACTACATCAGGTTTAGCTATGAGAAATCACATTCCAGGCCAGCAGAGAGGTCCTGAGGGATTCTGGCAACAGATATGGCTTTAAACATGCACTACTCACCAGGAGGAAGTTGGGTCAGGTAAGGGCTTCGCAGCCCCCCTGAGCCACTGATTCCTGTGTGCAGAAAGCCAAGAAGAAGAAGATTGAAGTGGTCAAGGCTGGAGACCTGGGCGT ...................................................................................................................................(((((((((((..((.(((.(((((...(((((....))))).))))).)))))..)))))).))).)).................................................. ...............................................................................................................................128.....................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|