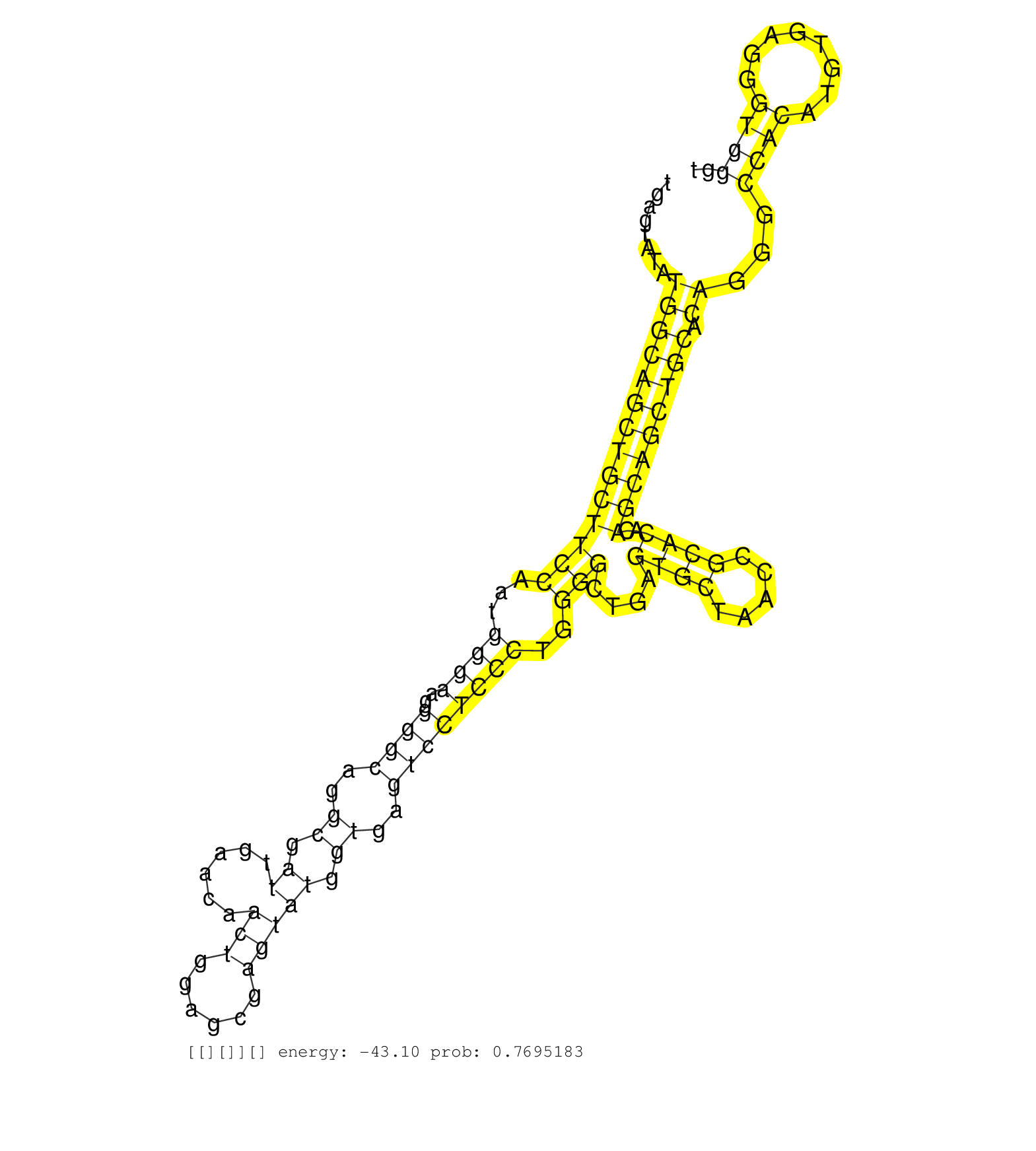

| Gene: AK049289 | ID: uc009gmf.1_intron_2_0_chr7_50078114_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| CTGCTGGATGCTGAGAGGGAAATTCAGGCGAGCTCTGACTTCGCGACATGGTGAGTATATGGCAGCTGCTTCCAATGGGAAGGGGCAGGCGATTGAACAACTGGAGCGAGTATGGTGAGTCCTCCCTGGGGCTGAGTGCTAACCGCACACAGCAGCTGCACAGGGCCACATGTGAGGGTGGGTGTGGTCTTAGCTGCTCCCAGAGAAAGCATGAAGGTGACCTCTGAATAACTTTGCTCTGGGGCTGCAT ...........................................................((((((((((((((...((((..((((..((.((......(((......))))).))..))))))))..)))....((((.....))))..))))))))).))...((((........))))..................................................................... ...................................................52.................................................................................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................CTCCCTGGGGCTGAGTGCTAACCGCACACAGCAGCTGCACAGGGCCACATGT............................................................................. | 52 | 2 | 36.00 | 36.00 | 36.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGCTGGATGCTGAGAGGGAAATTCAGGC............................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GAATAACTTTGCTCTctc....... | 18 | ctc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTGAACAACTGGAGCGAGTATGGgg..................................................................................................................................... | 25 | gg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGAGAGGGAAATTCAGGCGAGCTCTGA.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGGATGCTGAGAGGGAAATTCAGGC............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTGAACAACTGGAGCGAGTATGGatgc................................................................................................................................... | 27 | atgc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................ATATGGCAGCTGCTgtga................................................................................................................................................................................ | 18 | gtga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................CAGGCGAGCTCTGACTTagc.............................................................................................................................................................................................................. | 20 | agc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTGAACAACTGGAGCGAGTATGGTG..................................................................................................................................... | 25 | 7 | 0.71 | 0.71 | - | - | - | 0.29 | - | - | - | - | - | - | 0.14 | - | - | - | 0.14 | 0.14 | - | - | - | - |
| .........................AGGCGAGCTCTGACT.................................................................................................................................................................................................................. | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| .............................................................................................TGAACAACTGGAGCGAGTATGGTGAG................................................................................................................................... | 26 | 7 | 0.43 | 0.43 | - | 0.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGAACAACTGGAGCGAGTATGGTGA.................................................................................................................................... | 25 | 7 | 0.43 | 0.43 | - | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - |
| .............................................................................................TGAACAACTGGAGCGAGTATGGTGAGTC................................................................................................................................. | 28 | 6 | 0.33 | 0.33 | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - |
| .....................ATTCAGGCGAGCTCTGA.................................................................................................................................................................................................................... | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - |
| ............................................................................................TTGAACAACTGGAGCGAGTATGGT...................................................................................................................................... | 24 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - |
| ...........................................................................................ATTGAACAACTGGAGCGAGTATGG....................................................................................................................................... | 24 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ATTGAACAACTGGAGCGAGTATGGT...................................................................................................................................... | 25 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ATTGAACAACTGGAGCGAGTATGGTG..................................................................................................................................... | 26 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - |
| .....................................................................................CAGGCGATTGAACAACTGGAGCGAGTA.......................................................................................................................................... | 27 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |
| ...................................................................................GGCAGGCGATTGAACAACTGGAGCGAGT........................................................................................................................................... | 28 | 8 | 0.12 | 0.12 | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGCGATTGAACAACTGGAGCGAGTAT......................................................................................................................................... | 26 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - |
| ...........................................................................................ATTGAACAACTGGAGCGAG............................................................................................................................................ | 19 | 8 | 0.12 | 0.12 | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................AGGCGATTGAACAACTGGAGCGAGTA.......................................................................................................................................... | 26 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - |
| ...........................................................................TGGGAAGGGGCAGGCGATTGAACAAC..................................................................................................................................................... | 26 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - |
| ..................................................GTGAGTATATGGCAGCTGCTTCCAATG............................................................................................................................................................................. | 27 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 |
| CTGCTGGATGCTGAGAGGGAAATTCAGGCGAGCTCTGACTTCGCGACATGGTGAGTATATGGCAGCTGCTTCCAATGGGAAGGGGCAGGCGATTGAACAACTGGAGCGAGTATGGTGAGTCCTCCCTGGGGCTGAGTGCTAACCGCACACAGCAGCTGCACAGGGCCACATGTGAGGGTGGGTGTGGTCTTAGCTGCTCCCAGAGAAAGCATGAAGGTGACCTCTGAATAACTTTGCTCTGGGGCTGCAT ...........................................................((((((((((((((...((((..((((..((.((......(((......))))).))..))))))))..)))....((((.....))))..))))))))).))...((((........))))..................................................................... ...................................................52.................................................................................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................AGTGCTAACCGtcgg......................................................................................................... | 15 | tcgg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................ATGGTGAGTATATGGCAGCTGCTTCCA................................................................................................................................................................................ | 27 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 | - | - | - | - | - | - | - |