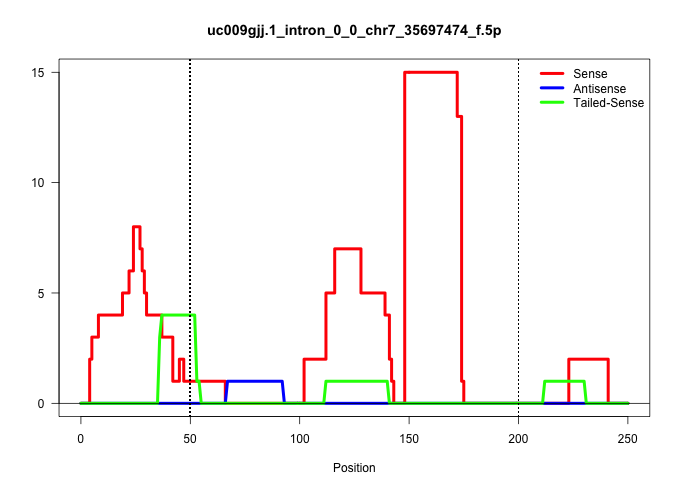
| Gene: Pepd | ID: uc009gjj.1_intron_0_0_chr7_35697474_f.5p | SPECIES: mm9 |
 |
|
(7) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| CTGCACTCCACGTGACGCTAGCGCCGAGTGAACATGGCGTCCACTGTGCGGTGAGTGCTGAGCTTGTCCCTTGCCCTGCCTGCGGCCCGCGTAGAGGTGGCCTGCCTCGGTATTCCTAGCTGAACAGTGGGTGACAGCCTCCGAGCTCTGACTGGAGTTGCTCTGTGGCCGCGGGCTGCGGGCTGCGCGCTTCTTCTCTGGGGCACCGGAACCGAGAGCCGGGCTGGGCGAGTGGTACCTGCAGAGCCCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TGACTGGAGTTGCTCTGTGGCCGCGG............................................................................ | 26 | 1 | 12.00 | 12.00 | 2.00 | 2.00 | - | 5.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................GCGTCCACTGTGCGGTt..................................................................................................................................................................................................... | 17 | t | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTCCTAGCTGAACAGTGGGTGACAGCCTC............................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................CGAGTGAACATGGCGTCC................................................................................................................................................................................................................ | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGACTGGAGTTGCTCTGTGGCCGC.............................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................................CTGGGCGAGTGGTACCTG......... | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGCCTCGGTATTCCTAGCTGAACAGT.......................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTCCTAGCTGAACAGTGGGTGACAGCCTa............................................................................................................. | 29 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................CGTCCACTGTGCGGTatc................................................................................................................................................................................................... | 18 | atc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....CTCCACGTGACGCTAGCGCCGAG.............................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CACGTGACGCTAGCGCCGAGTG............................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................GCCGAGTGAACATGGCGTCCACTGT........................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....ACTCCACGTGACGCTAGCGCCGA............................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGACTGGAGTTGCTCTGTGGCCGCGGG........................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ACTCCACGTGACGCTAGCGCCGAGT............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CGAGAGCCGGGCTGGacgc................... | 19 | acgc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................GTGCGGTGAGTGCTGAGCTTG........................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................TAGCTGAACAGTGGGTGACAGCCTCC............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGCTGAACAGTGGGTGACAGCCTCCG........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................AGCGCCGAGTGAACATGG..................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTCCTAGCTGAACAGTGGGTGACAGCC............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGCACTCCACGTGACGCTAGCGCCGAGTGAACATGGCGTCCACTGTGCGGTGAGTGCTGAGCTTGTCCCTTGCCCTGCCTGCGGCCCGCGTAGAGGTGGCCTGCCTCGGTATTCCTAGCTGAACAGTGGGTGACAGCCTCCGAGCTCTGACTGGAGTTGCTCTGTGGCCGCGGGCTGCGGGCTGCGCGCTTCTTCTCTGGGGCACCGGAACCGAGAGCCGGGCTGGGCGAGTGGTACCTGCAGAGCCCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................CCCTTGCCCTGCCTGCGGCCCGCGTA............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |