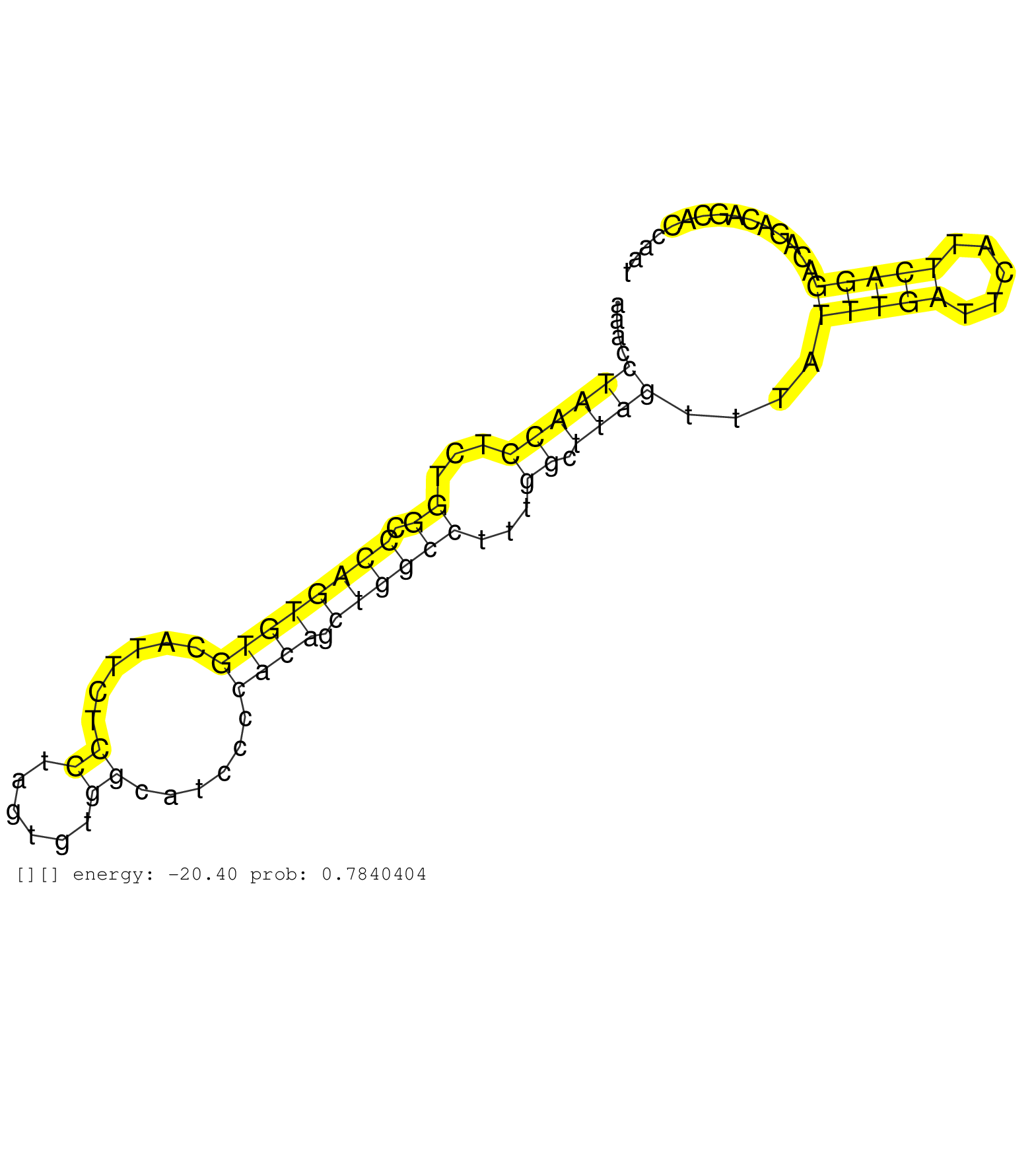

| Gene: Zbtb32 | ID: uc009gfh.1_intron_4_0_chr7_31376985_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| CCCTAAACCCACTATCTGATACCTCTGCTAACTGCCTGGCTGTCGTTCTTCAGGCCCCAAACCTAACCTCTGGCCCAGTGTGCATTCTCCTAGTGTGGCATCCCCACAGCTGGCCTTTGGCTTAGTTTATTTGATTCATTCAGGACAGACAGCACCAATCTGAGCCCAGACACTCACTCTTTCTCCCTTCCCCTTCACAGGCCAGAGAGTATGCGGTTTCTCTCCCACTTCAATCATGGATCCCCCATTG ..............................................................((((((...((.((((((((......((......))......)))).))))))...)).))))....(((((.....))))).......................................................................................................... ..........................................................59..................................................................................................159......................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TATTTGATTCATTCAGGACAGACAGCAC............................................................................................... | 28 | 1 | 10.00 | 10.00 | 6.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TATTTGATTCATTCAGGACAGACAGCA................................................................................................ | 27 | 1 | 6.00 | 6.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ...............................................................................................................................TATTTGATTCATTCAGGACAGACAGCACC.............................................................................................. | 29 | 1 | 5.00 | 5.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................TAACCTCTGGCCCAGTGTGCATTCTCC................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TATTTGATTCATTCAGGACAGACAGC................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTCATTCAGGACAGACAGCACCAATCT......................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................TTGGCTTAGTTTATTTGATTCATTCAGGA......................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................CTAACCTCTGGCCCAGTGTGCATTCTC................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TTTGATTCATTCAGGACAGACAGCACC.............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................TTGGCTTAGTTTATTTGATTCATTCAG........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................TGATTCATTCAGGACAGACAGCACCAATt.......................................................................................... | 29 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTCATTCAGGACAGACAGCACCAATC.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................TGATTCATTCAGGACAGACAGCACCAAT........................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................ATTTGATTCATTCAGGACAGACAGCAC............................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................TCCCCTTCACAGGCCAGAGAGTATGCGG.................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................TTAGTTTATTTGATTCATTCAGGACA....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................ATTCATTCAGGACAGACAGCACCAATCT......................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TTGGCTTAGTTTATTTGATTCATTCAGGACA....................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................TCTGAGCCCAGACACTCACTCTTTCTCCC............................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................TGCTAACTGCCTGGCTGTCGTTCTTC....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TAACCTCTGGCCCAGTGTGCATTCTtcta.............................................................................................................................................................. | 29 | tcta | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....AACCCACTATCTGATACCTCTGCTAACTGC....................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CCCTAAACCCACTATCTGATACCTCTGCTAACTGCCTGGCTGTCGTTCTTCAGGCCCCAAACCTAACCTCTGGCCCAGTGTGCATTCTCCTAGTGTGGCATCCCCACAGCTGGCCTTTGGCTTAGTTTATTTGATTCATTCAGGACAGACAGCACCAATCTGAGCCCAGACACTCACTCTTTCTCCCTTCCCCTTCACAGGCCAGAGAGTATGCGGTTTCTCTCCCACTTCAATCATGGATCCCCCATTG ..............................................................((((((...((.((((((((......((......))......)))).))))))...)).))))....(((((.....))))).......................................................................................................... ..........................................................59..................................................................................................159......................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................CCTCTGGCCCAGTGTGCAT..................................................................................................................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CCTCTGGCCCAGTGTGCATTCTCCTA.............................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GCTGGCCTTTGGCTTAGTTTATTTGATTCA................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................ACTCTTTCTCCCTTCCCCTTCACA................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GCCCAGTGTGCATTCTCCTAGTGTGGCA...................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TCCCCTTCACAGGCCAGAGAGTATGCG................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GCTAACTGCCTGGCTGTCGTTCTTCA...................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................CTGGCCCAGTGTGCATTCTCCTAGTG........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGCTAACTGCCTGGCTGTCGTTCTTCA...................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |