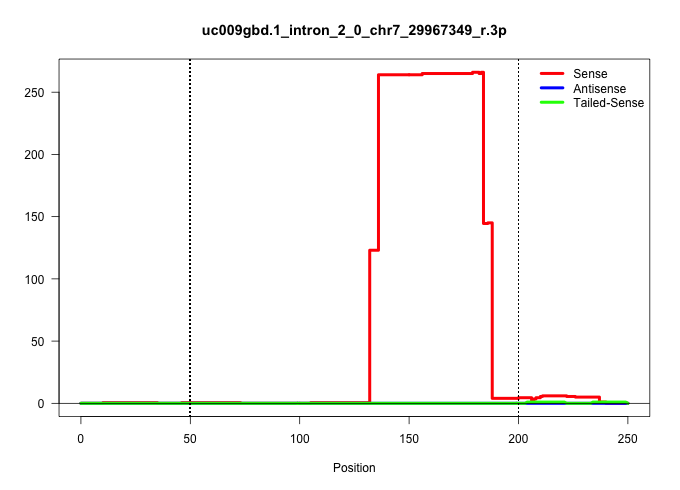
| Gene: A230107C01Rik | ID: uc009gbd.1_intron_2_0_chr7_29967349_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(12) TESTES |
| GGTGTGGCTCTGTTGGAGGGTGCTTGCCTGCAGTACTGCAATGAGTTGAGTCTTGGCCCAGCAGTAGAATGCTTGCCTAACTCTCATGAGGCCCCAAGTTCAGTCTGAAGCACCATAGTCACCAAGCAGCAGGCAGATTGGAGAAATTAGTGTTTCCCCTGGACTGTAGGTGCTACAGCTGTCTACTTTTGGACTTGTAGGTGGCTGTGCTTGGAGAACTCCCCTTGCTATGACATCATCCCCCAAAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................ATTGGAGAAATTAGTGTTTCCCCTGGACTGTAGGTGCTACAGCTGTCTACTT.............................................................. | 52 | 1 | 141.00 | 141.00 | 141.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................GCAGATTGGAGAAATTAGTGTTTCCCCTGGACTGTAGGTGCTACAGCTGTCT.................................................................. | 52 | 1 | 123.00 | 123.00 | 123.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTGGAGAACTCCCCTTGCTATGACATC............. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................ACTTTTGGACTTGTAGGTGGCT............................................ | 22 | 2 | 1.50 | 1.50 | - | - | - | 1.50 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CCCTGGACTGTAGGTGCTACAGCTGT.................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGGAGAACTCCCCTTGCTATGACATCATC.......... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................ATCATCCCCCAAtatg | 16 | tatg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................TGCTTGGAGAACTCCCCTTGCTATGACATC............. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGTCTACTTTTGGACTTGTAGGTGGCT............................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CTGTGCTTGGAGAACgaa............................ | 18 | gaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................TACTTTTGGACTTGTAGGTGGCTGTGC........................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GCTTGGAGAACTCCCCTTGCTATGACATC............. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGCTTGGAGAACTCCCCTT........................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ........................................................................................................................................................................................................GTGGCTGTGCTTGGAGAACTCC............................ | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..........TGTTGGAGGGTGCTTGCCTGCAGTA....................................................................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..........................................................................................................................................................................................TTTTGGACTTGTAGGTGGCTGTGCT....................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..............................................TGAGTCTTGGCCCAGCAGTAGAATGCT................................................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAAGCACCATAGTCACCAAGCAGCAG...................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| GGTGTGGCTCTGTTGGAGGGTGCTTGCCTGCAGTACTGCAATGAGTTGAGTCTTGGCCCAGCAGTAGAATGCTTGCCTAACTCTCATGAGGCCCCAAGTTCAGTCTGAAGCACCATAGTCACCAAGCAGCAGGCAGATTGGAGAAATTAGTGTTTCCCCTGGACTGTAGGTGCTACAGCTGTCTACTTTTGGACTTGTAGGTGGCTGTGCTTGGAGAACTCCCCTTGCTATGACATCATCCCCCAAAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................GCCCAGCAGTAGAAggca..................................................................................................................................................................................... | 18 | ggca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |