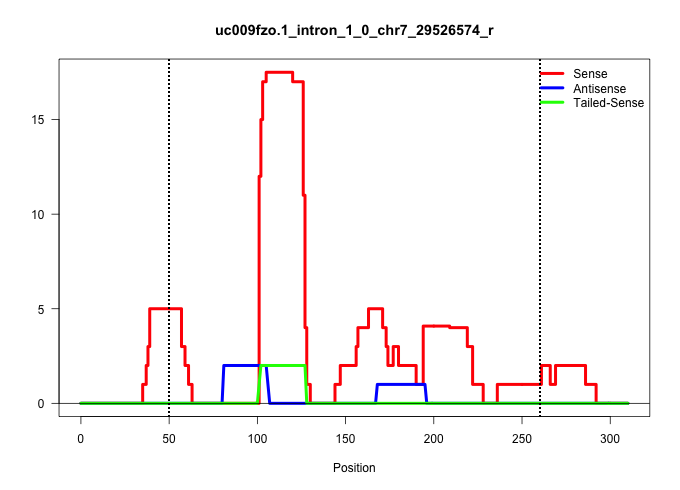
| Gene: Mrps12 | ID: uc009fzo.1_intron_1_0_chr7_29526574_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| GGGGCTGGAGGTAGGGGTTGGCGTGTGTGCGACCCCGCTAGGTTGGTGAGGTGGGTGCAGTTATCTGTGATCCAGATCTCTCCTCTCTCCTCAGTTTCCCCTTCCAGTATCCGGGCAAGGGCGGTTGGGTCCTTGGGGGCCTACTCAGAAGGAGACTGAATTTGCGAGGGGACTTTCTGTTTTAGAGGAGATTCTAGAAGGCTCTGAGAGTGGGTTTCTGGTCCCAACCTCCTTTCTGAATGTCTTCTCTCCTAATACAGGGACGCTGCAGGCGGCGAGATGTCCTGGCCCGGCCTTCTTTATGGCCTCA ................................................................................................(((((((((........)).))))).))..(((.((..((..((((((((...((....))..........((((.(((((((((((....)))))...))))))))))..))))))))..)))).)))..................................................................................... ................................................................................................97...............................................................................................................................226.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesWT1() Testes Data. (testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TTCCAGTATCCGGGCAAGGGCGGTT........................................................................................................................................................................................ | 25 | 1 | 6.00 | 6.00 | - | - | 2.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TTCCAGTATCCGGGCAAGGGCGGTTG....................................................................................................................................................................................... | 26 | 1 | 6.00 | 6.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................AGGTTGGTGAGGTGGGTG............................................................................................................................................................................................................................................................. | 18 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................TCCAGTATCCGGGCAAGGGCGGTTGG...................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TCAGAAGGAGACTGAATTTGCGAGGG............................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TAGAAGGCTCTGAGAGTGGGTTTCTGGT........................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCCAGTATCCGGGCAAGGGCGGTTGt...................................................................................................................................................................................... | 26 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTTTTAGAGGAGATTCTAGAAGGCTC.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................TGAATGTCTTCTCTCCTAATACAGGGACGC............................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................AGGTTGGTGAGGTGGGTGCAGT......................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCCAGTATCCGGGCAAGGGCGGTTG....................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................GACGCTGCAGGCGGCGAGATGTCCTGGCCCG.................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................TAGGTTGGTGAGGTGGGTGCAGTTA....................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GAAGGAGACTGAATTTGCGAGGGGACT........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCGAGGGGACTTTCTGTTTTAGAGGAG........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TTCCAGTATCCGGGCAAGGGCGGTTGt...................................................................................................................................................................................... | 27 | t | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AGGCGGCGAGATGTCCT........................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TCAGAAGGAGACTGAATTTGCGAGGGG........................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CGCTAGGTTGGTGAGGTGGGTGCA........................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................CTAGGTTGGTGAGGTGGGTG............................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TAGAAGGCTCTGAGAGTGGGTTTCT........................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAATTTGCGAGGGGACTTTCTGT.................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CCAGTATCCGGGCAAGGGCGGTTGGGT.................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TCCTTTCTGAATGTCTTCTCTCCTAAT...................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................GAATTTGCGAGGGGAC......................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CCAGTATCCGGGCAAGGGCGGTTGG...................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAGAGTGGGTTTCTGGTCCCAAC.................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTGGGTGCAGTTATCTG................................................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................................................................................AGTATCCGGGCAAGG.............................................................................................................................................................................................. | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATCTGTGATCCAGATCTC...................................................................................................................................................................................................................................... | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ..................................................................................................................................................................................................TAGAAGGCTCTGAGA..................................................................................................... | 15 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - |
| GGGGCTGGAGGTAGGGGTTGGCGTGTGTGCGACCCCGCTAGGTTGGTGAGGTGGGTGCAGTTATCTGTGATCCAGATCTCTCCTCTCTCCTCAGTTTCCCCTTCCAGTATCCGGGCAAGGGCGGTTGGGTCCTTGGGGGCCTACTCAGAAGGAGACTGAATTTGCGAGGGGACTTTCTGTTTTAGAGGAGATTCTAGAAGGCTCTGAGAGTGGGTTTCTGGTCCCAACCTCCTTTCTGAATGTCTTCTCTCCTAATACAGGGACGCTGCAGGCGGCGAGATGTCCTGGCCCGGCCTTCTTTATGGCCTCA ................................................................................................(((((((((........)).))))).))..(((.((..((..((((((((...((....))..........((((.(((((((((((....)))))...))))))))))..))))))))..)))).)))..................................................................................... ................................................................................................97...............................................................................................................................226.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesWT1() Testes Data. (testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................TCCCAACCTCCTTTCTGA....................................................................... | 18 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CCTCTCTCCTCAGTTTCCCCTTCCA............................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................GGGACTTTCTGTTTTAGAGGAGATTCTA.................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................CCTCTCTCCTCAGTTTCCCCTTCCAG........................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |