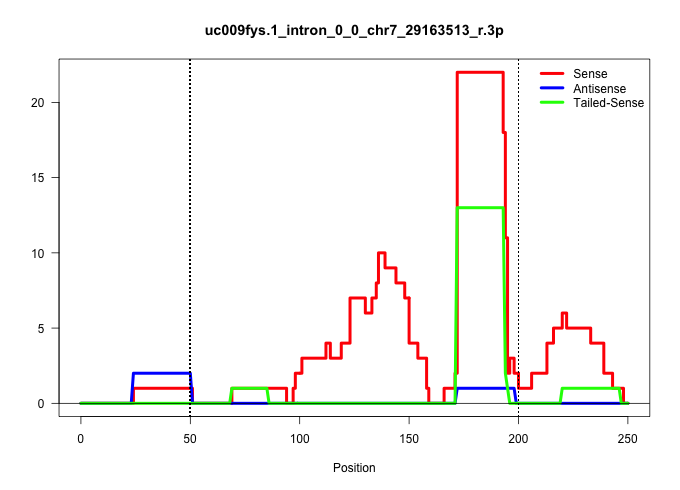
| Gene: Zfp36 | ID: uc009fys.1_intron_0_0_chr7_29163513_r.3p | SPECIES: mm9 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| TCCTTCTGGATTACCTTCAGTCATGTGGAAACACTCAAATCTCTGTAGTCAGGTTCTCCCTGGAGTTTGTGTGAGCCGGGTGGGGCTCAGGCCGAGAACCCACAAACCGGACTGGCAAGCTCGTGAAGTGCTTTACAACAGGGGCGGGGCGACGTCACCTTACCTTTTCTGGTGGCCTAACCGAACCCTGTTCCTCTCAGAGCCTCCAGTCGATGAGCCATGACCTGTCATCCGACCACGGAGGAACCGA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTCt........................................................ | 22 | t | 11.00 | 4.00 | 6.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTCCT....................................................... | 23 | 1 | 9.00 | 9.00 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTCC........................................................ | 22 | 1 | 7.00 | 7.00 | - | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTC......................................................... | 21 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................TGAAGTGCTTTACAACAGGGGCGGGGC.................................................................................................... | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGAGCCATGACCTGTCATCCGACCAC........... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................AACAGGGGCGGGGCGACGTCAC............................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................TGGCAAGCTCGTGAAGTGCTTTACAAC............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................TTCTGGTGGCCTAACCGAACCCTGTTCCTCTCAG.................................................. | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTCtt....................................................... | 23 | tt | 1.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAACAGGGGCGGGGCGACGTCACC........................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGAGCCGGGTGGGac.................................................................................................................................................................... | 17 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CAGTCGATGAGCCATGACCTGTCATCC................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................ACAAACCGGACTGGCAAGCTCGTGAAGTG........................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACAACAGGGGCGGGGCGACG................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................TGTGAGCCGGGTGGGGCTCAGGCCG............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTattc...................................................... | 24 | attc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GTGGAAACACTCAAATCTCTGTAGTCA....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CTCGTGAAGTGCTTTACAACAGGGG.......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGAAGTGCTTTACAACAGGGGCGGG...................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CCCACAAACCGGACTGGCAAGCTCG............................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................GTGGCCTAACCGAACCCTGTTCCTCTC.................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGAGCCTCCAGTCGATGAGCCATG............................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................ACCCACAAACCGGACTG........................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGACCTGTCATCCGACCACGGAGGAACC.. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGACCTGTCATCCGACCACGGAGGAcc... | 27 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GCCATGACCTGTCATCCGACCACGGAG....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCTTCTGGATTACCTTCAGTCATGTGGAAACACTCAAATCTCTGTAGTCAGGTTCTCCCTGGAGTTTGTGTGAGCCGGGTGGGGCTCAGGCCGAGAACCCACAAACCGGACTGGCAAGCTCGTGAAGTGCTTTACAACAGGGGCGGGGCGACGTCACCTTACCTTTTCTGGTGGCCTAACCGAACCCTGTTCCTCTCAGAGCCTCCAGTCGATGAGCCATGACCTGTCATCCGACCACGGAGGAACCGA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................GTGGAAACACTCAAATCTCTGTAGTCA....................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GAGAACCCACAAACCGGACTGGCAaaa..................................................................................................................................... | 27 | aaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGCCTAACCGAACCCTGTTCCTCTCA................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |