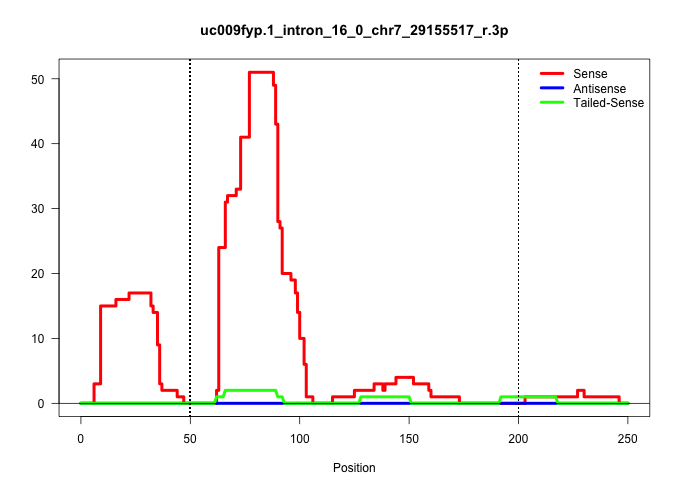
| Gene: Plekhg2 | ID: uc009fyp.1_intron_16_0_chr7_29155517_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| TGTGGTTGATGGAAGATGTGAATAAAAGCACTGGAGGTTTAGGTCCCAGAGAGGAAGGAGCATTGATAGGTACTGAATCAGACTGGAGAAGTAGAGGAGTGGAGCAGAGACCTGTGGGGACTGTCACCTGAGTGTGGGTGACCATGGGGACCGTAAGACCGTGTGTCTGGGAAGCCCTGACCACTTGCTCCCTACCTCAGCAGCGCAGGCAGCCACCACCATGGCTTCTCCAAGGGGTTCTGGCAGCTCC ...........................................................((....((((((.(((..((..(((.....)))....))......)))..))))))..((((.(((((((......)))))))(((((...........))))).)))).....))........................................................................... ..........................................................59....................................................................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................TGATAGGTACTGAATCAGACTGGAGAA................................................................................................................................................................ | 27 | 1 | 15.00 | 15.00 | 1.00 | 1.00 | 3.00 | 3.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................TCAGACTGGAGAAGTAGAGGAGTGGA................................................................................................................................................... | 26 | 1 | 8.00 | 8.00 | 2.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAGGTACTGAATCAGACTGGAGAAGT.............................................................................................................................................................. | 26 | 1 | 7.00 | 7.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TCAGACTGGAGAAGTAGAGGAGTGG.................................................................................................................................................... | 25 | 1 | 7.00 | 7.00 | 2.00 | 2.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGGAAGATGTGAATAAAAGCACTGGAG...................................................................................................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | - | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGGAAGATGTGAATAAAAGCACTGGA....................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | - | 1.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGATAGGTACTGAATCAGACTGGAGA................................................................................................................................................................. | 26 | 1 | 5.00 | 5.00 | 1.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAATCAGACTGGAGAAGTAGAGGAGT...................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAATCAGACTGGAGAAGTAGAGGAG....................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGATGGAAGATGTGAATAAAAGCACT.......................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................CTGAATCAGACTGGAGAAGTAGAG.......................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGGGGACCGTAAGACCGTGTGTCTGGGAA............................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGTGACCATGGGGACCGTAAGACC.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........TGGAAGATGTGAATAAAAGCACTGGAGG..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TTGATAGGTACTGAATCAGACTGGAGA................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................ACTGAATCAGACTGGAGAAGTAGAGGA........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TCAGACTGGAGAAGTAGAGGAGTGGAG.................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GGGGACTGTCACCTGAGTGTGGG................................................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................CTCCAAGGGGTTCTGGCAG.... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................TGATAGGTACTGAATCAGACTGGAGAAG............................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TCAGACTGGAGAAGTAGAGGAGTGGAGCA................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGAGTGTGGGTGACCATGGGGcc................................................................................................... | 23 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................TGGGTGACCATGGGGACCGTAAGACC.......................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TCAGACTGGAGAAGTAGAGGAGTGGAGC................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................TGATAGGTACTGAATCAGACTGGAG.................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGGTACTGAATCAGACTGGAGAAGTAGAG.......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................TGAATCAGACTGGAGAAGTAGAGGA........................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................TGTGAATAAAAGCACTGGAGGTTTAGGT.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAGGTACTGAATCAGACTGGAGAAGTt............................................................................................................................................................. | 27 | t | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................TACCTCAGCAGCGCAGGCAGCCccca................................ | 26 | ccca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......TGATGGAAGATGTGAATAAAAGCACTG......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................ACCTGAGTGTGGGTGACCATGGGGACC.................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................TTGATAGGTACTGAATCAGACTGGcgaa................................................................................................................................................................ | 28 | cgaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TTGATAGGTACTGAATCAGACTGGAG.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................ACTGAATCAGACTGGAGAAG............................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................CGCAGGCAGCCACCACCATGGCTTCTC.................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TAAAAGCACTGGAGGTTTAGGTCCC........................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GACCATGGGGACCGTAAGAC........................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTGGTTGATGGAAGATGTGAATAAAAGCACTGGAGGTTTAGGTCCCAGAGAGGAAGGAGCATTGATAGGTACTGAATCAGACTGGAGAAGTAGAGGAGTGGAGCAGAGACCTGTGGGGACTGTCACCTGAGTGTGGGTGACCATGGGGACCGTAAGACCGTGTGTCTGGGAAGCCCTGACCACTTGCTCCCTACCTCAGCAGCGCAGGCAGCCACCACCATGGCTTCTCCAAGGGGTTCTGGCAGCTCC ...........................................................((....((((((.(((..((..(((.....)))....))......)))..))))))..((((.(((((((......)))))))(((((...........))))).)))).....))........................................................................... ..........................................................59....................................................................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|