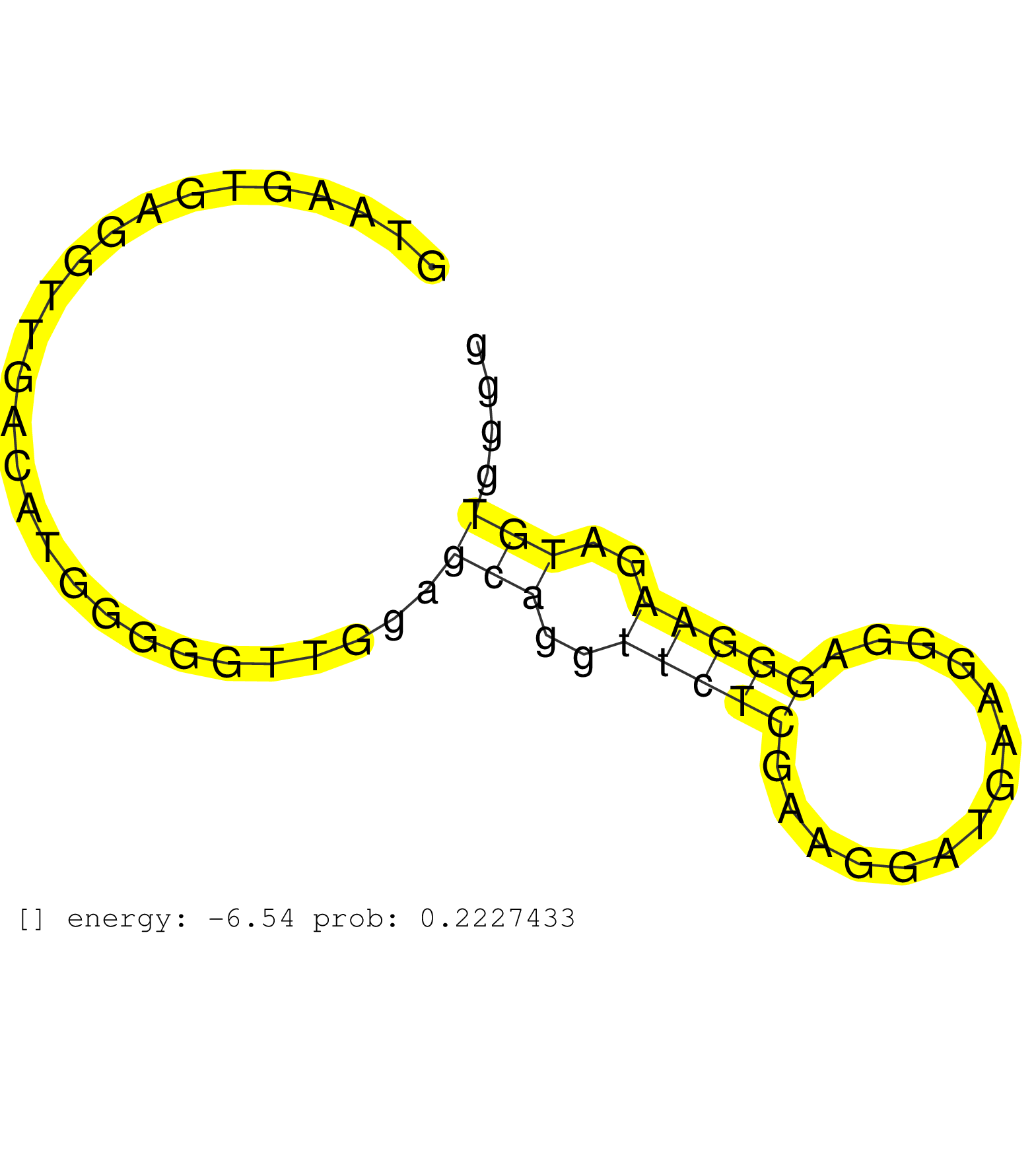

| Gene: Fbl | ID: uc009fxy.1_intron_1_0_chr7_28959706_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(3) PIWI.mut |
(19) TESTES |
| AGGCGGAGGGGGTGGTGGCTTCAGGGGACGAGGAGGTGGCGGTGGCCGAGGTAAGTGAGGTTGACATGGGGGTTGGAGCAGGTTCTCGAAGGATGAAGGGAGGGAAGATGTGGGGGCCTCCCTCTCCTACTGCAGCTCTCTCTTACAGGTGGGGGCTTCCAGTCTGGGGGCAACCGGGGTCGAGGTGGTGGCCGGGGA .............................................................................(((..(((((..............)))))..)))....................................................................................... ..................................................51..............................................................115................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGAGGTTGACATGGGGGTTG........................................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| ...............................................................................................................................TACTGCAGCTCTCTCTTACAGt................................................. | 22 | t | 4.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TCGAAGGATGAAGGGAGGGAAGATGT....................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGAGGTTGACATGGGGG.............................................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CCTACTGCAGCTCTCTCTTACAG.................................................. | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGAGGTTGACATGGGGGTT............................................................................................................................ | 24 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TCAGGGGACGAGGAGGTGGCGGgg.......................................................................................................................................................... | 24 | gg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TCTCGAAGGATGAAGGGAGGGAAGA.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GCAACCGGGGTCGAaga............ | 17 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................TCTCGAAGGATGAAGGGAGGGAAG........................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTGAGGTTGACATGGGGGTTG........................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GACGAGGAGGTGGagag........................................................................................................................................................... | 17 | agag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CCTACTGCAGCTCTCTCTTACAGaa................................................ | 25 | aa | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................CTACTGCAGCTCTCTCTTACAGaaaa.............................................. | 26 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGTGAGGTTGACATGGGGGTTGGA......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GACATGGGGGTTGGAGCAGGTTCT................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGAAGGGAGGGAAGATGTGGGGGC................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGAGGTTGACATGGGGGT............................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................CGAAGGATGAAGGGAGGGAAGATGT....................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTGAGGTTGACATGGGGGTTGGA......................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TTCAGGGGACGAGGAGGTGGCGGa........................................................................................................................................................... | 24 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGAGGTTGACATGGGGGTTG........................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................GGCAACCGGGGTCGAGGTGGTG........ | 22 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TTCCAGTCTGGGGGCAACCGGGGT.................. | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| .......................GGGGACGAGGAGGTGGCGG............................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .............................................................................................................................................................TCCAGTCTGGGGGCAACCGG..................... | 20 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GTGGTGGCTTCAGGGGACGAGGAG................................................................................................................................................................... | 24 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GTGGTGGCTTCAGGGGACGA....................................................................................................................................................................... | 20 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CAGGGGACGAGGAGGTGG............................................................................................................................................................... | 18 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGCGGAGGGGGTGGTGGCTTCAGGGGACGAGGAGGTGGCGGTGGCCGAGGTAAGTGAGGTTGACATGGGGGTTGGAGCAGGTTCTCGAAGGATGAAGGGAGGGAAGATGTGGGGGCCTCCCTCTCCTACTGCAGCTCTCTCTTACAGGTGGGGGCTTCCAGTCTGGGGGCAACCGGGGTCGAGGTGGTGGCCGGGGA .............................................................................(((..(((((..............)))))..)))....................................................................................... ..................................................51..............................................................115................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................CAGTCTGGGGGCAACCGGGGTCGAt............... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |