
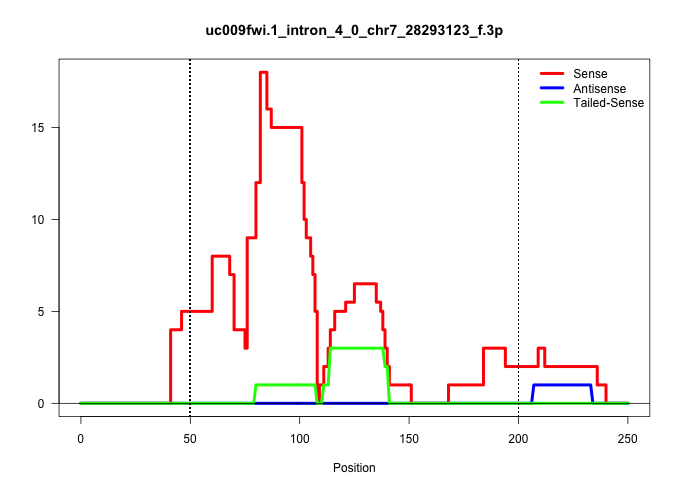
| Gene: Prx | ID: uc009fwi.1_intron_4_0_chr7_28293123_f.3p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| TCCGGAAAGGGGCGGGGCTAAAGCTAAAGCTCTTGGAGCCATAACCTCTGAAATTACCTCTCAAGCTAGAGGCCTGTGGATATGCAGGGTCAAAATCTGGGATTGGGCCTCTCTTGGCTGACTCCTTGGAAGTGGGTGGGCCCAAGTGTGGGTGGAGTCAGGAGACACTCACTCCAGACCCACCTCCACCTCGTTTGCAGGGGACCAGCTGCTGAGTGCCCGTGTGTTCTTTGAGAACTTCAAATATGAG ........................................................(((((......)))))(((((......))))).................................................................................................................................................................. ..................................................51....................................................105............................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................TGGATATGCAGGGTCAAAATCTGGG..................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGCAGGGTCAAAATCTGGGATTGGGC.............................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................TAACCTCTGAAATTACCTCTCAAGCTAGA.................................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ............................................................TCAAGCTAGAGGCCTGTGGATATGC..................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGGATATGCAGGGTCAAAATCTGGGA.................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGGCTGACTCCTTGGAAGTGGGTGGGt............................................................................................................. | 27 | t | 2.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TATGCAGGGTCAAAATCTGGGATTGGGC.............................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TCCACCTCGTTTGCAGGGGACCAGCTGC...................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGCAGGGTCAAAATCTGGGATTGGG............................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................TCTCTTGGCTGACTCCTTGGAAGTGG................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGCAGGGTCAAAATCTGGGATTGG................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................TATGCAGGGTCAAAATCTGGGATTG................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................................TGCTGAGTGCCCGTGTGTTCTTTGAGA.............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................TATGCAGGGTCAAAATCTGGGATTGGGt.............................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GCTGACTCCTTGGAAGTGGGTGGGC............................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TCAAGCTAGAGGCCTGTGGATATGCAG................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................TTGGCTGACTCCTTGGAAGTGGGTGG............................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................TTGGAAGTGGGTGGGCCCAAGTGTGG................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGGATATGCAGGGTCAAAATCTGGGAT................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCTTGGCTGACTCCTTGGAAGTGGGTGt............................................................................................................... | 28 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCTTGGCTGACTCCTTGGAAGTGGGTG................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TCTGAAATTACCTCTCAAGCTAGAGGCCT............................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TAACCTCTGAAATTACCTCTCAAGCTA...................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGAGTGCCCGTGTGTTCTTTGAGAACTT.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TCACTCCAGACCCACCTCCACCTCGT........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................TGGCTGACTCCTTGGAAGTGGGTGGG.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................CTCCTTGGAAGTGGGT................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| TCCGGAAAGGGGCGGGGCTAAAGCTAAAGCTCTTGGAGCCATAACCTCTGAAATTACCTCTCAAGCTAGAGGCCTGTGGATATGCAGGGTCAAAATCTGGGATTGGGCCTCTCTTGGCTGACTCCTTGGAAGTGGGTGGGCCCAAGTGTGGGTGGAGTCAGGAGACACTCACTCCAGACCCACCTCCACCTCGTTTGCAGGGGACCAGCTGCTGAGTGCCCGTGTGTTCTTTGAGAACTTCAAATATGAG ........................................................(((((......)))))(((((......))))).................................................................................................................................................................. ..................................................51....................................................105............................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................GCTGCTGAGTGCCCGTGTGTTCTTTGA................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |