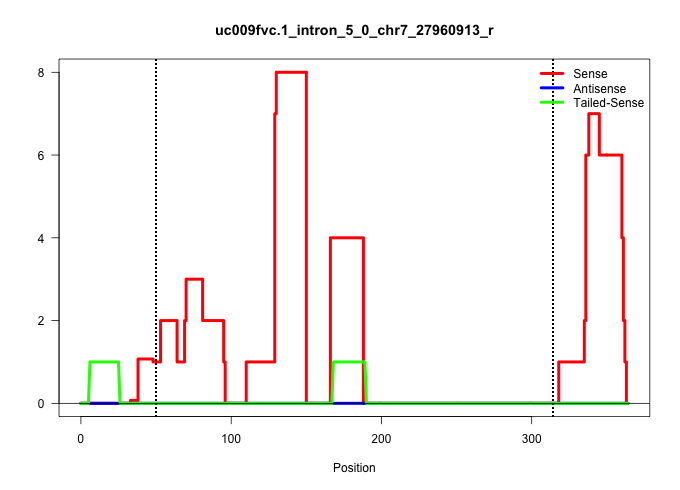
| Gene: Rab4b | ID: uc009fvc.1_intron_5_0_chr7_27960913_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(11) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(23) TESTES |
| TGCTGGAACTGGCAAATCATGTCTTCTCCACCAGTTTATTGAGAATAAGTGTGAGTTTCTGGCAGGACCCTGGAACTGCGGATGTGGTCATGGTGTGGGTCAGTGGGGCAGCAGGTGGGGTAGGTAGCAGGTACCATTGACTAAAGCTAGTACTATAGGCTTGCCTCTAGCTTTGGTGGATGGTCTTTGCTAAGTTCTCTGCCAACCCAGCAGTGAGAATTGGGGTACAGAAAAGGAGGAAGAGGTCTGCAGGATGGGGGGGATTCCAGGAGCCCCTGCACTGCCCCCCCACCCCGTTTCCCTCCTACTCTCAGTCAAACAGGACTCCAACCACACTATCGGCGTGGAGTTTGGATCCAGGGTG .....................................................(((..(((((...(((.((..(((((....))))))).)))...(((((((((.((.((..............))..)).)))))))))...))))).))).................................................................................................................................................................................................................. ................................................49.......................................................................................................154................................................................................................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................GGTACCATTGACTAAAGCTAG...................................................................................................................................................................................................................... | 21 | 1 | 10.00 | 10.00 | 1.00 | - | - | 2.00 | - | 1.00 | - | - | 1.00 | - | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................CTAGCTTTGGTGGATGGTCTTT................................................................................................................................................................................ | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| .................................................................................................................................GGTACCATTGACTAAAGCT........................................................................................................................................................................................................................ | 19 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................AAAAGGAGGAAGAGGTCTGCAGGATa............................................................................................................ | 26 | a | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................AAACAGGACTCCAACCACACTATCGGCGT................... | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CAGGTGGGGTAGGTAGCA........................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................TATCGGCGTGGAGTTTGGATCCAG.... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................TGGTCATGGTGTGGGTCAGTGGGGCAGCAGGTG....................................................................................................................................................................................................................................................... | 33 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AACTGGCAAATCATGTCatg.................................................................................................................................................................................................................................................................................................................................................. | 20 | atg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TTGGTGGATGGTCTTTGCTAAG.......................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................GGTACCATTGACTAAAGCTA....................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GCAGGTGGGGTAGGTAGCA........................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AGGTACCATTGACTAAAGCTAGa..................................................................................................................................................................................................................... | 23 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................TATCGGCGTGGAGTTTGGATCCAGG... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................TCAAACAGGACTCCAACCACACTATCGGCGT................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................AGCTTTGGTGGATGGTCTTTGt.............................................................................................................................................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................TATCGGCGTGGAGTTTGGATCCAGGG.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................ACAGGACTCCAACCACACTATCGGCGT................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGGAACTGCGGATGTGGTCATGGTGT............................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................AGCTTTGGTGGATGGTCTTTGa.............................................................................................................................................................................. | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGGAACTGCGGATGTGGTCATGGTG............................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................................................................................................................................ACACTATCGGCGTGGAGTTTGGA......... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTACCATTGACTAAAGCTAG...................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TTGAGAATAAGTGTGAGTTTCTGGCA............................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTTTCTGGCAGGACCCTGGAACTGCGG........................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................TCGGCGTGGAGTTTGGATCCAGGGT. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................CTATCGGCGTGGAGTTTGGATCCAGG... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................GTTTATTGAGAATAA............................................................................................................................................................................................................................................................................................................................ | 15 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - |
| TGCTGGAACTGGCAAATCATGTCTTCTCCACCAGTTTATTGAGAATAAGTGTGAGTTTCTGGCAGGACCCTGGAACTGCGGATGTGGTCATGGTGTGGGTCAGTGGGGCAGCAGGTGGGGTAGGTAGCAGGTACCATTGACTAAAGCTAGTACTATAGGCTTGCCTCTAGCTTTGGTGGATGGTCTTTGCTAAGTTCTCTGCCAACCCAGCAGTGAGAATTGGGGTACAGAAAAGGAGGAAGAGGTCTGCAGGATGGGGGGGATTCCAGGAGCCCCTGCACTGCCCCCCCACCCCGTTTCCCTCCTACTCTCAGTCAAACAGGACTCCAACCACACTATCGGCGTGGAGTTTGGATCCAGGGTG .....................................................(((..(((((...(((.((..(((((....))))))).)))...(((((((((.((.((..............))..)).)))))))))...))))).))).................................................................................................................................................................................................................. ................................................49.......................................................................................................154................................................................................................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................GTAGCAGGTACCATtcg................................................................................................................................................................................................................................... | 17 | tcg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |