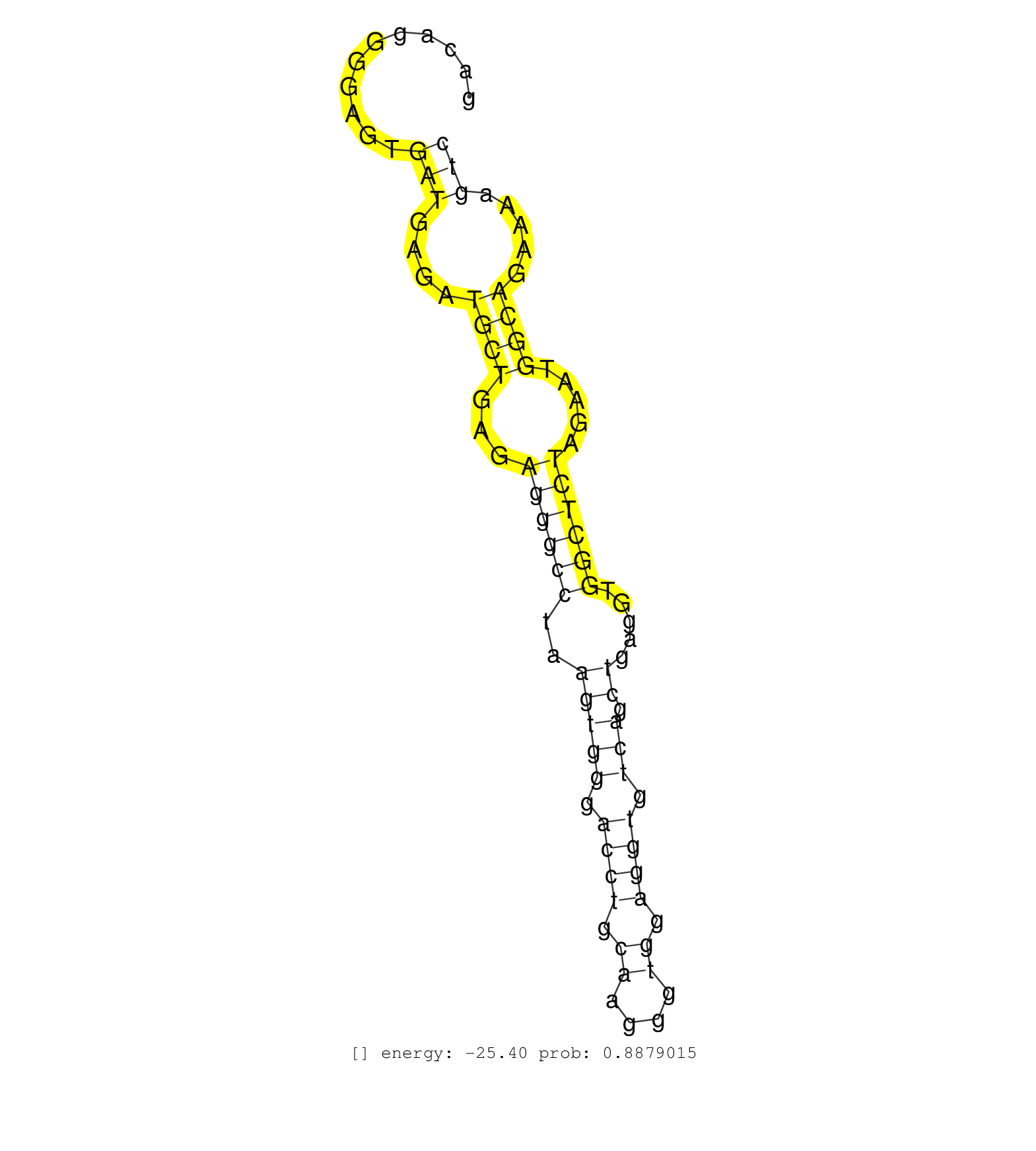
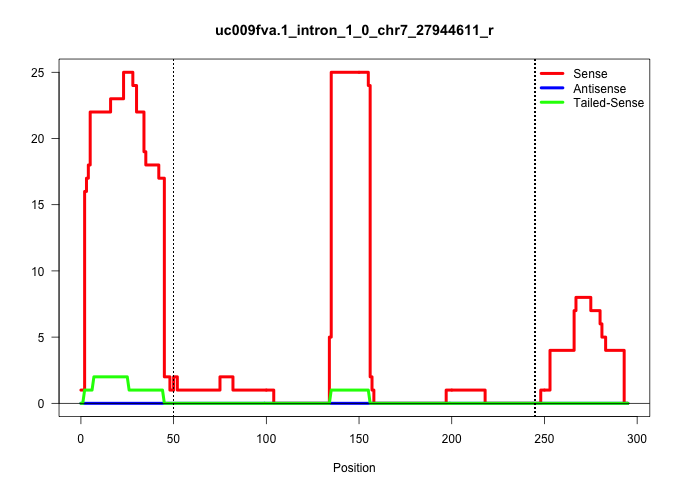
| Gene: Egln2 | ID: uc009fva.1_intron_1_0_chr7_27944611_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(1) PIWI.mut |
(29) TESTES |
| TGGTCTGACCGACGGAACCCACATGAGGTGAAGCCAGCCTATGCCACCAGGTATGACCTGTACTTTAGGGATTGTCCAGCTGCTTCCCCTGTGCCAAACATTTCTTAGCTTGTCCCCAGTCCCCAGAGTTGACAGGGGAGTGATGAGATGCTGAGAGGGCCTAAGTGGGACCTGCAAGGGTGGAGGTGTCAGCTGAGGTGGCTCTAGAATGGCAGAAAAGTCATTCTGACTGCCCAATTTTTCAGGTACGCCATCACTGTCTGGTATTTTGATGCCAAGGAACGGGCAGCAGCCA .............................................................................................................................................(((....((((...((((((..(((((.((((.((....)).)))).))).)).....)))))).....)))).....)))......................................................................... ..................................................................................................................................131........................................................................................222....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................GGGAGTGATGAGATGCTGAGA........................................................................................................................................... | 21 | 1 | 18.00 | 18.00 | - | 5.00 | 3.00 | 1.00 | - | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................GGGAGTGATGAGATGCTGAGAGG......................................................................................................................................... | 23 | 1 | 6.00 | 6.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GGGGAGTGATGAGATGCTGAGA........................................................................................................................................... | 22 | 1 | 6.00 | 6.00 | - | - | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................TGAGGTGGCTCTAGAATGGCAG................................................................................ | 22 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACCGACGGAACCCACATGAGGTGAAGC..................................................................................................................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................................................................................TTTTGATGCCAAGGAACGGGCAGCAGC.. | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................ACTGTCTGGTATTTTGATG..................... | 19 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TCTGACCGACGGAACCCACATGAGGTG......................................................................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCACTGTCTGGTATTTTGATGCCAAGG............... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......ACCGACGGAACCCACATGt............................................................................................................................................................................................................................................................................. | 19 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACCGACGGAACCCACATGAGGTGA........................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................CGCCATCACTGTCTGGTATTTTGATGC.................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CCAGCTGCTTCCCCTGTGCCAAACATTTC............................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCACTGTCTGGTATTTTGATGCCAAGGA.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................GTGGCTCTAGAATGGCAGAAA............................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................TGAGGTGAAGCCAGCCTATGCCACCAGGT................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAGGTGAAGCCAGCCTATGCCACC....................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................ACCCACATGAGGTGAAGCCAGCCTAT............................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGAGTGATGAGATGCTGAG............................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCACTGTCTGGTATTTTGATGCCAAGGAAC............ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGAGTGATGAGATGCTGAGAG.......................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................CGCCATCACTGTCTGGTATTTTGATGt.................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....TGACCGACGGAACCCACATGAGGTGAAGCC.................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGACCTGTACTTTAGGGATTGTCCAGCTG..................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................TTTGATGCCAAGGAACGGGCAGCAGC.. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....CTGACCGACGGAACCCACATGAGGTG......................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGAGTGATGAGATGCTGAta........................................................................................................................................... | 21 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGGTCTGACCGACGGAACCCACATGAGGTGAAGCCAGCCTATGCCACCAGGTATGACCTGTACTTTAGGGATTGTCCAGCTGCTTCCCCTGTGCCAAACATTTCTTAGCTTGTCCCCAGTCCCCAGAGTTGACAGGGGAGTGATGAGATGCTGAGAGGGCCTAAGTGGGACCTGCAAGGGTGGAGGTGTCAGCTGAGGTGGCTCTAGAATGGCAGAAAAGTCATTCTGACTGCCCAATTTTTCAGGTACGCCATCACTGTCTGGTATTTTGATGCCAAGGAACGGGCAGCAGCCA .............................................................................................................................................(((....((((...((((((..(((((.((((.((....)).)))).))).)).....)))))).....)))).....)))......................................................................... ..................................................................................................................................131........................................................................................222....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GACCTGTACTTTAGGGATTGTCCAGCTGCTT.................................................................................................................................................................................................................. | 31 | 1 | 4.00 | 4.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |