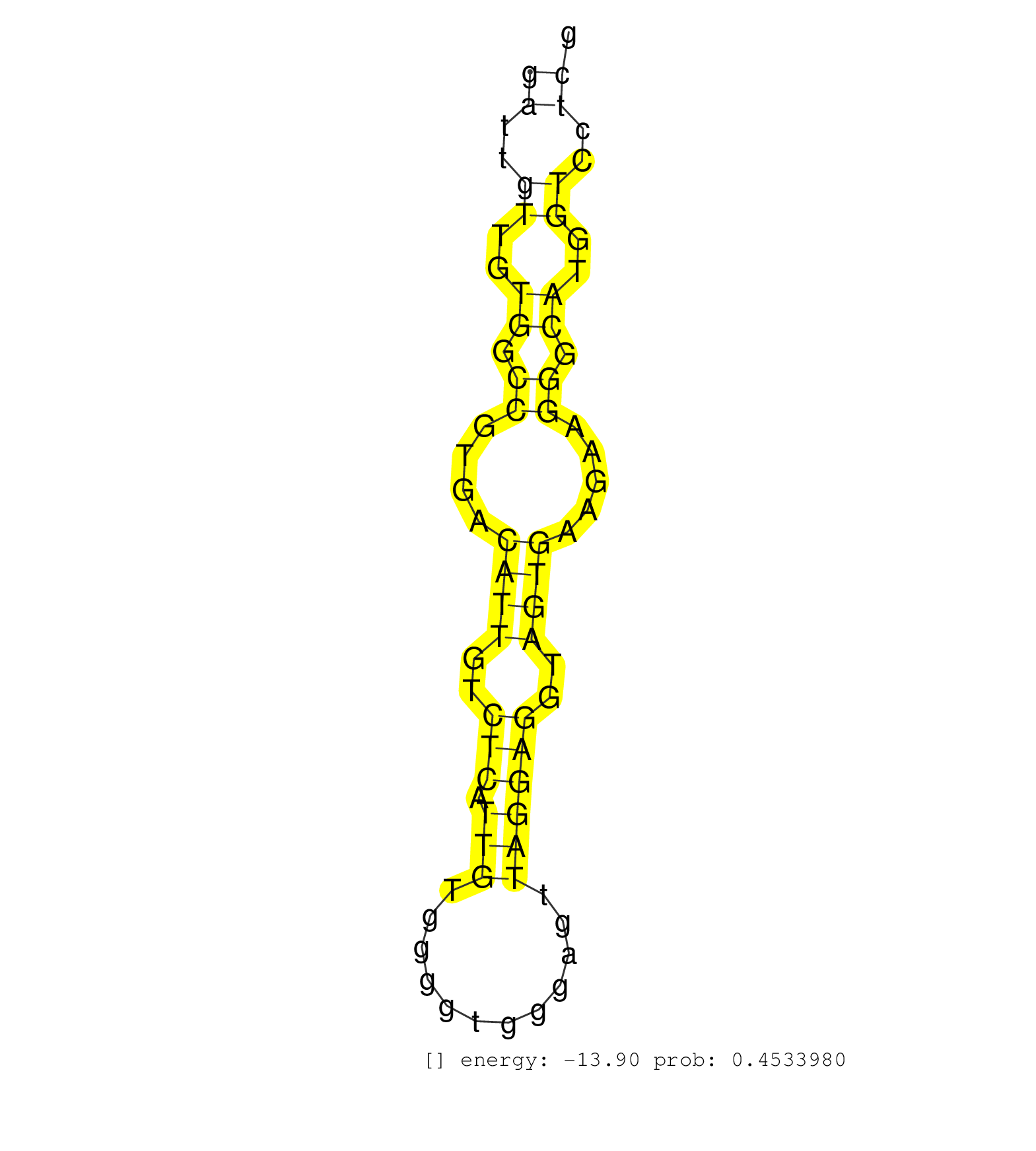

| Gene: Exosc5 | ID: uc009fti.1_intron_4_0_chr7_26451362_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| CATATAAAGCGAGGTGTAGTGACACTCTTAGGAGGCTGGGCAGGATGATTGTTGTGGCCGTGACATTGTCTCATTGTGGGGTGGGAGTTAGGAGGTAGTGAAGAAGGGCATGGTCCTCGTGATGGTGAGGAGGAGTAGAAGGGGAGGGAAGGAGGGAGGCTTAGGGATTCCTTTGGGGCTAACCCTCTCCTGGTTCCCAGCTCCAACAGTGTCTGGCCGCCGCCCAAGCCGCCTCACAGCACATCTTCCG ..............................................((..((..((.((....((((..(((.(((............))))))..)))).....)).))..))..)).................................................................................................................................... ..............................................47......................................................................119................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGG......................................................................................................................................... | 25 | 1 | 9.00 | 9.00 | 3.00 | - | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGGTC....................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | 2.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCGTGATGGTGAGGAGGAGTAGAAG............................................................................................................. | 25 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCGTGATGGTGAGGAGGAGTAGAAGG............................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCGTGATGGTGAGGAGGAGTAGAA.............................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGGTCCT..................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGGT........................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................TCGTGATGGTGAGGAGGAGTAGAAGt............................................................................................................ | 26 | t | 2.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TAGGAGGCTGGGCAGGATGATTGTTG.................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................TAGTGACACTCTTAGGAGGCTGGGCA................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TCTTAGGAGGCTGGGCAGGATGATTG....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CACTCTTAGGAGGCTGGGCAGGATG........................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGGTCCTCGTGATGGTGAGGAGGAGT.................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................TCTCCTGGTTCCCAGCTCCAACAGTGTC..................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGGTCCTC.................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGTAGTGACACTCTTAGGAGGCTGGGCAG............................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGGTCC...................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................TGACATTGTCTCATTGTGGGGTGGGAG................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CCAACAGTGTCTGGCCGCCGCCCAAGC..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATGGTCtt..................................................................................................................................... | 29 | tt | 1.00 | 6.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTGTGGCCGTGACATTGTCTCATTGT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AGGAGGTAGTGAAGAAGGGCATGGTCCTC.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AAGCGAGGTGTAGTGACACTCTTAGGAGGC...................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..TATAAAGCGAGGTGTAGTGACACTCT.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................CTGGGCAGGATGATTGTTGTGGCCGg............................................................................................................................................................................................. | 26 | g | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GAGTTAGGAGGTAGTGAAGAAGGGCATG.......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................TCGTGATGGTGAGGAGGAGTAGAAt............................................................................................................. | 25 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CCGCCCAAGCCGCCTCACAGCACATCTTC.. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCAT........................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TAAAGCGAGGTGTAGTGACACTCTT............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGGAGGTAGTGAAGAAGGGCATG.......................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CATATAAAGCGAGGTGTAGTGACACTCTTAGGAGGCTGGGCAGGATGATTGTTGTGGCCGTGACATTGTCTCATTGTGGGGTGGGAGTTAGGAGGTAGTGAAGAAGGGCATGGTCCTCGTGATGGTGAGGAGGAGTAGAAGGGGAGGGAAGGAGGGAGGCTTAGGGATTCCTTTGGGGCTAACCCTCTCCTGGTTCCCAGCTCCAACAGTGTCTGGCCGCCGCCCAAGCCGCCTCACAGCACATCTTCCG ..............................................((..((..((.((....((((..(((.(((............))))))..)))).....)).))..))..)).................................................................................................................................... ..............................................47......................................................................119................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................TCCTGGTTCCCAGCTC............................................... | 16 | 8 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - |