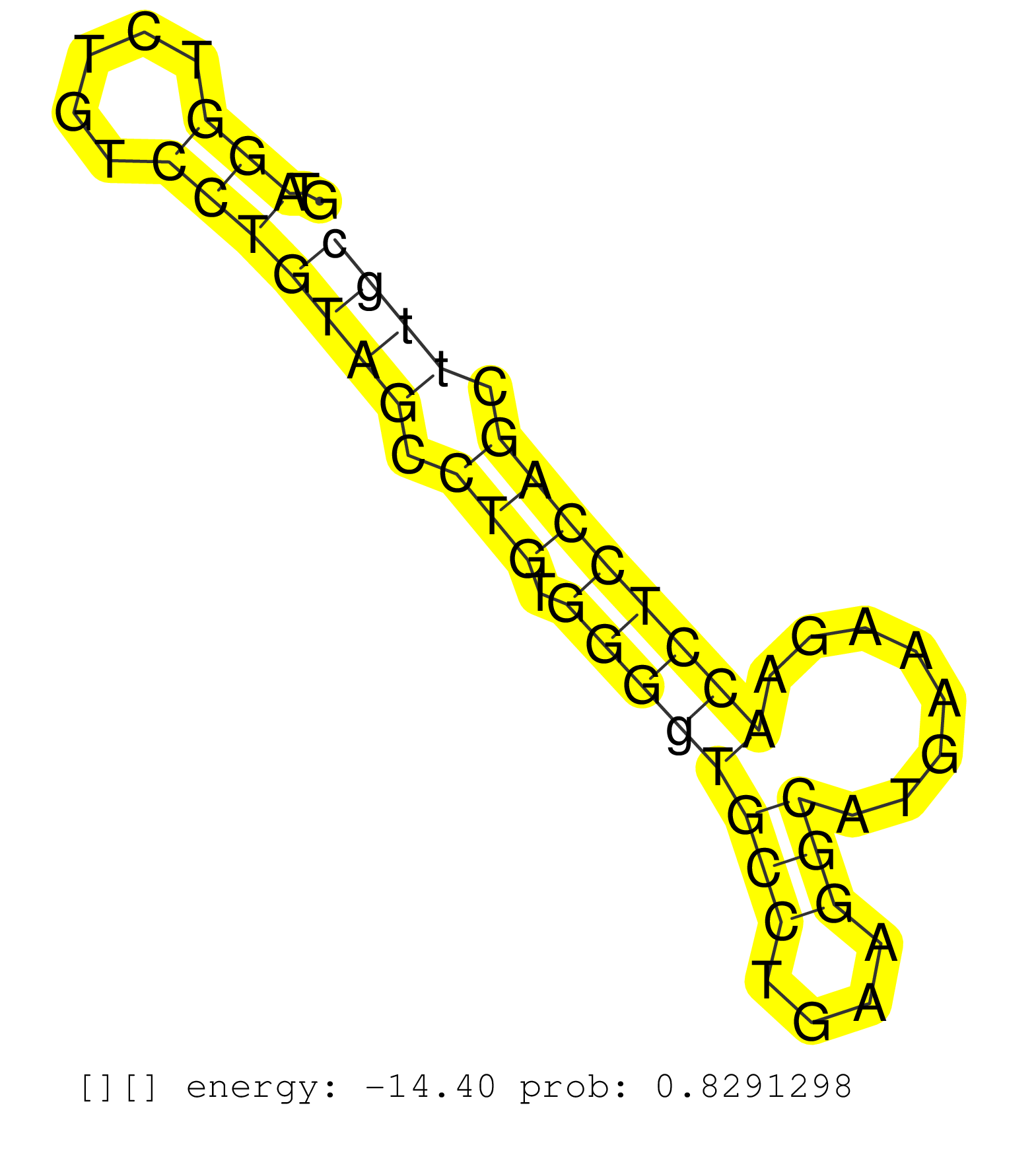
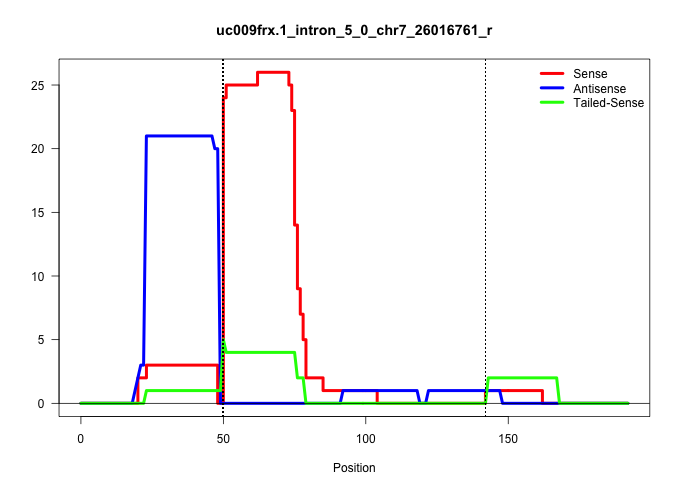
| Gene: Gsk3a | ID: uc009frx.1_intron_5_0_chr7_26016761_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| TTGCTTGTGGACCCTGACACTGCTGTCCTCAAGCTCTGCGATTTTGGCAGGTAGGTCTGTCCTGTAGCCTGTGGGGTGCCTGAAGGCATGAAAGAACCTCCAGCTTGCTTCCATGCTTAACCCTGCCTGGCTCTTCCCACAGTGCAAAGCAGCTGGTTCGGGGGGAGCCCAATGTGTCCTACATCTGCTCTC ....................................................(((.....)))((((.(((.((((((((....)))........)))))))).)))).................................................................................... ..................................................51.......................................................108.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGG..................................................................................................................... | 25 | 1 | 12.00 | 12.00 | 4.00 | - | - | 2.00 | 1.00 | - | - | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGGG.................................................................................................................... | 26 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGGGTG.................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGGGTGt................................................................................................................. | 29 | t | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGG...................................................................................................................... | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGGGTGC................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGCTGTCCTCAAGCTCTGCGATTTTGGC................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGGGT................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTGGtg.................................................................................................................... | 26 | tg | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGCAAAGCAGCTGGTTCGGG.............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TAGGTCTGTCCTGTAGCCTGTGGGGTGC................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTG....................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................TGTCCTCAAGCTCTGCGATTTTGGCAGt............................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGTAGCCTGTGGGGTGCCTGAAG........................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGCCTGAAGGCATGAAAGAACCTCCAGC........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GCAAAGCAGCTGGTTCGGGGGGAtt........................ | 25 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GCAAAGCAGCTGGTTCGGGGGGggc........................ | 25 | ggc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGCTGTCCTCAAGCTCTGCGA....................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................TGTCCTCAAGCTCTGCGATTTTGGC................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TCAAGCTCTGCGATTTTGGCAGGTAG.......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTGTCCTGTAGCCTGTtggg.................................................................................................................... | 26 | tggg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ACTGCTGTCCTCAAGCTCTGCGA....................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TTGTGGACCCTGACACTGCTGTCC.................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTGCTTGTGGACCCTGACACTGCTGTCCTCAAGCTCTGCGATTTTGGCAGGTAGGTCTGTCCTGTAGCCTGTGGGGTGCCTGAAGGCATGAAAGAACCTCCAGCTTGCTTCCATGCTTAACCCTGCCTGGCTCTTCCCACAGTGCAAAGCAGCTGGTTCGGGGGGAGCCCAATGTGTCCTACATCTGCTCTC ....................................................(((.....)))((((.(((.((((((((....)))........)))))))).)))).................................................................................... ..................................................51.......................................................108.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................TGTCCTCAAGCTCTGCGATTTTGGCA............................................................................................................................................... | 26 | 1 | 18.00 | 18.00 | - | 5.00 | 5.00 | - | - | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................CTGCCTGGCTCTTCCCACAGTGCAAA............................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................AGAACCTCCAGCTTGCTTCCATGCTTA......................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GCTGTCCTCAAGCTCTGCGATTTTGGCA............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GCCCAATGTGTCCTACATCTGCTCc.. | 25 | c | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGCTGTCCTCAAGCTCTGCGATTTTGGCA............................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGCTGTCCTCAAGCTCTGCGATTTTGG................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GCCTGGCTCTTCCCACAGTGCAAAGCA......................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |