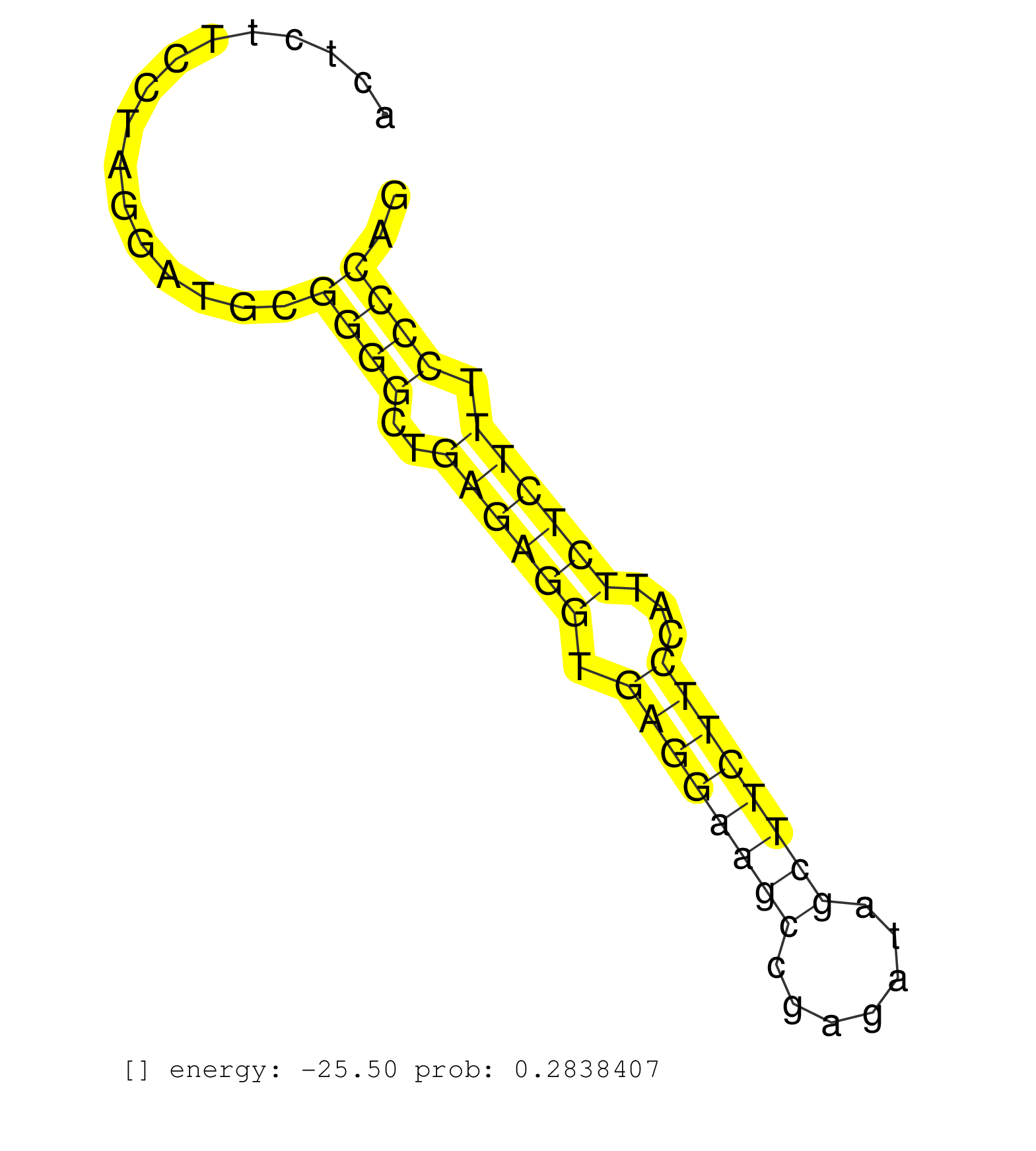
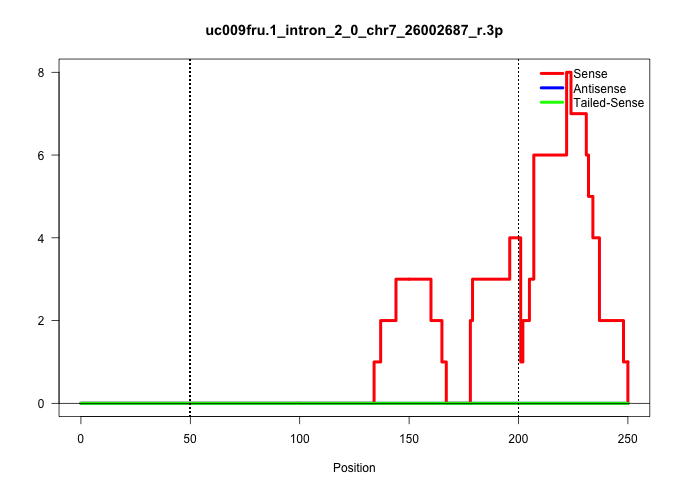
| Gene: Dedd2 | ID: uc009fru.1_intron_2_0_chr7_26002687_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| CGGGTTGAACCGCCCCATGAGTACTCTGTCCTGGGAGGCCTTTGAACAACCTCTGGGCGGGAGGTGCTTGAGGAAGCGTGGGTATTTGGGTGTGACCTCTGAGCGGCTGGGTCAGGCATCCCGTGGCTGGGCACTCTTCCTAGGATGCGGGGCTGAGAGGTGAGGAAGCCGAGATAGCTTCTTCCATTCTCTTTCCCCAGTGTCCCCTGAGCGATACAGCTATGGCAATCCCAGCTCTTCTTCCAAGAGG ....................................................................................................................................................((((..((((((.((((((((.......))))))))...)))))).)))).................................................... ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TTCTTCCATTCTCTTTCCCCAGT................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 |
| ...............................................................................................................................................................................................................TGAGCGATACAGCTATGGCAATCCCAGCTC............. | 30 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTTCCCCAGTGTCCCCTGAGCGATACAGC.............................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGCAATCCCAGCTCTTCTTCCAAGA.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................................CCAGTGTCCCCTGAGCGATACAGCTATG.......................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................................TGGCAATCCCAGCTCTTCTTCCAAGAGG | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTTCCATTCTCTTTCCCCAGT................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGAGCGATACAGCTATGGCAATCCCAG................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................TCCTAGGATGCGGGGCTGAGAGGTGAGG..................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCCCCTGAGCGATACAGCTATGGCAATCC................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TCTTCCTAGGATGCGGGGCTGAGAGG.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGCGGGGCTGAGAGGTGAGGAA................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CCTGAGCGATACAGCTATGGCAATCCC.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| CGGGTTGAACCGCCCCATGAGTACTCTGTCCTGGGAGGCCTTTGAACAACCTCTGGGCGGGAGGTGCTTGAGGAAGCGTGGGTATTTGGGTGTGACCTCTGAGCGGCTGGGTCAGGCATCCCGTGGCTGGGCACTCTTCCTAGGATGCGGGGCTGAGAGGTGAGGAAGCCGAGATAGCTTCTTCCATTCTCTTTCCCCAGTGTCCCCTGAGCGATACAGCTATGGCAATCCCAGCTCTTCTTCCAAGAGG ....................................................................................................................................................((((..((((((.((((((((.......))))))))...)))))).)))).................................................... ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................CCAGCTCTTCTTCCcatc...... | 18 | catc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |